Difference between revisions of "SOCR EduMaterials AnalysisActivities Fisher Exact"
m (→Example) |
(→Example) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 46: | Line 46: | ||
===Example=== | ===Example=== | ||
| − | * [http://www.r-project.org/ R calculations] | + | * If the “Cochran conditions” are satisfied (no cell has count zero, and more than 80% of the cells have counts at least 5) the [http://www.r-project.org/ R calculations] are based on asymptotic chi-squared probabilities and matrix simulation is done conditional on the row and column marginals, and works only if the marginals are strictly positive (Patefield algorithm, 1981). |
<code> > data <- matrix(c(13, 6, 29, 11, 16, 4), nr=3)</code><br> | <code> > data <- matrix(c(13, 6, 29, 11, 16, 4), nr=3)</code><br> | ||
<code> > data</code><br> | <code> > data</code><br> | ||
| Line 61: | Line 61: | ||
* SOCR Calculations | * SOCR Calculations | ||
<center>[[Image:SOCR_AnalysisActivities_FishersExactTest_Fig4.png|500px]]</center> | <center>[[Image:SOCR_AnalysisActivities_FishersExactTest_Fig4.png|500px]]</center> | ||
| − | + | ||
{{translate|pageName=http://wiki.stat.ucla.edu/socr/index.php?title=SOCR_EduMaterials_AnalysisActivities_FisherExact}} | {{translate|pageName=http://wiki.stat.ucla.edu/socr/index.php?title=SOCR_EduMaterials_AnalysisActivities_FisherExact}} | ||
Latest revision as of 15:40, 22 June 2010
Contents
[hide]SOCR Analysis Activities - SOCR Analyses Example on Fisher's Exact Test Activity
Applet
SOCR Fisher's exact Test Applet is available online here.
Background
You can find more details about the Fisher's Exact Test here. This demo is based on an example from Wolfram MathWorld Fisher's Exact Test.
Steps
1. Click on "Fisher's Exact Test" from the conbo box in the left panel.

2. Click on "Input" button. You'll see a screen like below waiting for your input

The minimum size of table is 2 by 2. We'll use 2 by 2 for our example. (Note that you can also change the number of rows and/or columns by clicking on the combo box.)
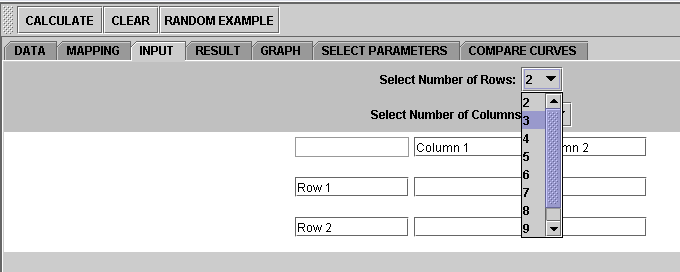
Type in numbers into the cells. You can also change the text of the row and column names from "Row 1" etc., to something you like.

3. Click on the "Calculate" button and then "Result" to see the results.
Note: if you happen to click on the "Clear" button in the middle of the procedure, all the data will be cleared out. Simply start over from step 1 and retrieve the data by clicking an EXAMPLE button.
Notes
- The SOCR Fisher's Exact Test provides calculations for examining the significance of the association (contingency) between the two kinds of classifications. These are in general represented by NxK matrices.
- Please note that in the following situations the Exact Fisher's test is very difficult to compute:
- anytime the frequency counts (cell-values in the matrix) are large (say >9), and/or
- the matrix-sizes > 2 (i.e., N and/or K >2).
- P-value-cutoff is \(p_o\), the starting likelihood of the matrix, which is used incrementally to compute the one-sided and the two-sided p-values of interest. Note that in general, the two-sided p-value will be quite different from 2 times the one-sided p-value!
- In SOCR Analyses, to make this test work for all these situations, we use approximate p-values calculations according to the MaskMetropolis MCMC method for random matrix generation.
- If your data is in any of these situations, we strongly recommend you also try the SOCR Chi-Square Contingency Table Test, which is more stable, albeit also approximate. See the SOCR Chi-Square Contingency Table Activity.
Example
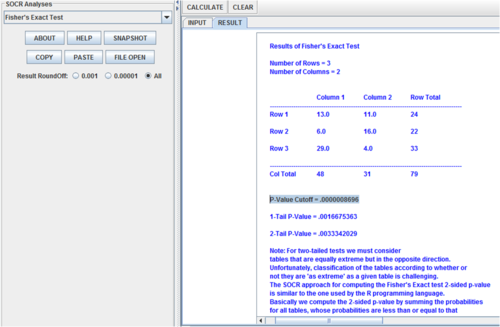
- If the “Cochran conditions” are satisfied (no cell has count zero, and more than 80% of the cells have counts at least 5) the R calculations are based on asymptotic chi-squared probabilities and matrix simulation is done conditional on the row and column marginals, and works only if the marginals are strictly positive (Patefield algorithm, 1981).
> data <- matrix(c(13, 6, 29, 11, 16, 4), nr=3)
> data
[,1] [,2]
[1,] 13 11
[2,] 6 16
[3,] 29 4
> fisher.test(data, alternative = "greater")
Fisher's Exact Test for Count Data
data: data
p-value = 0.00001568
alternative hypothesis: greater
- SOCR Calculations
Translate this page: