Difference between revisions of "SMHS DataManagement"
(→Theory) |
(→Theory) |
||
| Line 135: | Line 135: | ||
| − | + | <center> | |
| − | Dec Hex Binary | + | {|class="wikitable" style="text-align:center; width:33%" border="1" |
| − | + | |- | |
| − | + | |Dec||Hex||Binary | |
| − | + | |- | |
| − | + | |0||00||00000000 | |
| − | + | |- | |
| − | + | |1||01||00000001 | |
| − | + | |- | |
| − | + | |2||02||00000010 | |
| − | + | |- | |
| − | + | |3||03||00000011 | |
| − | + | |- | |
| − | + | |4||04||00000100 | |
| − | + | |- | |
| − | + | |5||05||00000101 | |
| − | + | |- | |
| − | + | |6||06||00000110 | |
| − | + | |- | |
| + | |7||07||00000111 | ||
| + | |- | ||
| + | |8||08||00001000 | ||
| + | |- | ||
| + | |9||09||00001001 | ||
| + | |- | ||
| + | |10||0A||00001010 | ||
| + | |- | ||
| + | |11||0B||00001011 | ||
| + | |- | ||
| + | |12||0C||00001100 | ||
| + | |- | ||
| + | |13||0D||00001101 | ||
| + | |- | ||
| + | |14||0E||00001110 | ||
| + | |- | ||
| + | |15||0F||00001111 | ||
| + | |- | ||
| + | |16||10||00010000 | ||
| + | |} | ||
| + | </center> | ||
Revision as of 15:37, 31 July 2014
Contents
Scientific Methods for Health Sciences - Data Management
Overview
Data management comprised all the discipline related to managing data as a valuable resource and it is of significant importance in various fields. It is officially defined as the development and execution of architectures, policies, practices and procedures that properly manage the full data lifecycle needs of an enterprise. There are various ways to manage data. In this lecture, we are going to introduce the fundamental roles of data management in statistics and illustrate commonly used ways and steps in data managements through examples from different areas including tables, streams, cloud, warehouses, DBs, arrays, binary ASC II, handling and mechanics.
Motivation
The next step after getting data would be to make proper management of the data in hand. Data management is of course a vital step in data analysis and is crucial to the success and reproducibility of a statistical analysis. So how would we do a good data management and what are the commonly used ways of data management? In order to further make good use of the data, we are going to learn more about the area of data management and various ways to implement it. Selection of appropriate tools and efficient use of these tools can save the researcher numerous hours, and allow other researchers to leverage the products of their work.
Theory
3.1) Tables: one of the most commonly used ways to manage data. It is a means of arranging data in rows and columns. It is of pervasive use throughout all research and data analysis. There are two basic types of tables:
- Simple Table: consider the following example of a table summarizing the data from two groups. The table presents a general comparison of the two groups and shows us a clear picture of the measurements and the comparative characteristics between the two groups. What we learn from the table below: (1) these two groups with the same sample size have the same mean; (2) group 1 has a bigger range of the data values; (3) group 2 has a smaller standard deviation indicating a less variant dataset compared to group 1.
| Minimum | Maximum | Mean | Standard Deviation | Size | |
| Group 1 | 12 | 45 | 22 | 2.6 | 40 |
| Group 2 | 15 | 30 | 22 | 1.5 | 40 |
- Multi-dimensional table: consider the F distribution (http://socr.umich.edu/Applets/F_Table.html) where the first row and the column is the degree of freedoms and the data in the middle is the 99% quantiles of F(k,l). This is a two dimensional example and the coordinates or combinations of the basic headers give a unique value attached.
| l \ k | 1 | 2 | 3 | 4 | 5 |
| 1 | 4052. | 4999.5 | 5403. | 5625. | 5764. |
| 2 | 98.50 | 99.00 | 99.17 | 99.25 | 99.30 |
| 3 | 34.12 | 30.82 | 29.46 | 28.71 | 28.24 |
| 4 | 21.20 | 18.00 | 16.69 | 15.98 | 15.52 |
| 5 | 13.27 | 12.06 | 11.39 | 10.97 | 10.67 |
3.2) Streams: is also an easy way of data management and it is more visualized with pictures. It is a sequence of data elements made available over time and can be thought as a conveyor belt that allows items to be processed one at a time rather than in large batches. It is a sequence of digitally encoded coherent signals used to transmit or receive information that is in the process of being transmitted. In electronics and computer architecture, it determines for which time which data item is scheduled to enter or leave which port.
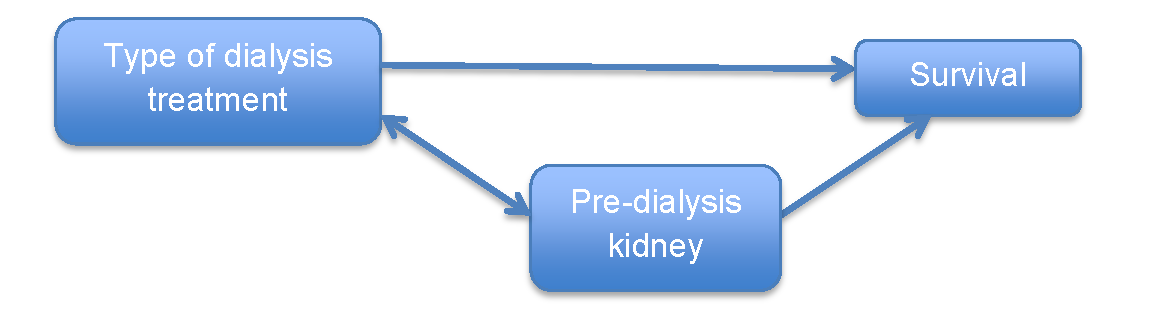
From the chart below, we have three apparent observations: (1) pre-dialysis kidney function is associated with patient survival; (2) pre-dialysis kidney function differs by type of dialysis treatment (it is associated with type of dialysis); (3) pre-dialysis kidney function is not in the causal pathway of type of dialysis and survival. And from these three conditions, we can conclude that pre-dialysis kidney function is a potential confounder.
An illustrative example of streaming is wind map (http://hint.fm/wind/), which generates a vivid demonstration of the wind speed over the USA with data streamed live from different resources. We can have a clear picture of the wind range countrywide with streaming.
3.3) Cloud Data Storage: Data cloud storage demands high availability, durability, and scalability from a few bytes to petabytes. Examples of data cloud storage services include Amazon’s S3, which promises a 99.9% monthly availability and 99.999999999% durability per year. This translates into less than an hour outage per month. For an example of durability, assume that a user stores 10,000 objects in a cloud storage, then, on average, the user would expect to lose one object every 10,000,000 years. S3, and other cloud service providers, achieve this reliability by storing data in multiple facilities with error checking and self-healing processes to detect and repair errors and device failures. This process is completely transparent to the user and requires no actions or knowledge of the underlying complexities. Global data-intense service providers like Google and Facebook have the expertise and scale to provide enormous storage and significant reliability and uptime in an efficient/economical way. Many Big Data research projects benefit from using cloud storage services (e.g., umich email, file-sharing, computational services etc.)
3.4) Warehouse: a system used for reporting and data analysis. Integrating data from one or more disparate sources creates a central repository of data, a data warehouse (DW). DW stores current and historical data and are used for creating trending reports for senior management reporting such as annual comparison.
3.5) DBs: database is an organized collection of data where data are typically organized to model relevant aspects of reality in a way that supports processes requiring this information.
A well-known example would be the pipeline, which utilizes data from the LONI Image Data Archive (IDA). The pipeline takes advantage of the cluster nodes to download files in parallel from the IDA database. (http://pipeline.loni.usc.edu/learn/user-guide/building-a-workflow/#IDA) This article (https://ida.loni.usc.edu/login.jsp) provides a specific introduction to The LONI Image Data Archive (IDA), which offers an integrated environment for safely archiving, querying, visualizing and sharing neuroimaging data. It facilitates the de-identification and pooling of data from multiple institutions, protecting data from unauthorized access while providing the ability to share data among collaborative investigators. The archive provides flexibility in establishing project metadata, accommodating one or more research groups, sits and others. It’s simple, secure and requires nothing more than a computer with Internet connection and a web browser.
3.6) Arrays: a data structure consisting of a collection of elements, each identified by at least one array index or key. Types of arrays:
- One-dimensional array: a simple example: an array of 8 integer variables with indices 0 through 7 may be sored as 8 words at memory address: {200, 202, 204, 206, 208, 210, 212, 214} which can be memorized as 200+2i.
- Multidimensional arrays: Data=[■(2&3&0@6&4&5@5&3&1)].
Example: R arrays and data-frames
DF <- data.frame(a = 1:3, b = letters[10:12],
c = seq(as.Date("2004-01-01"), by = "week", len = 3),
stringsAsFactors = TRUE)
> DF
a b c
1 1 j 2004-01-01 2 2 k 2004-01-08 3 3 l 2004-01-15
data.matrix(DF[1:2]) data.matrix(DF)
> data.matrix(DF)
a b c
[1,] 1 1 12418 [2,] 2 2 12425 [3,] 3 3 12432
> sleep # sleep dataset
extra group ID
1 0.7 1 1 2 -1.6 1 2 3 -0.2 1 3 4 -1.2 1 4 5 -0.1 1 5 6 3.4 1 6 7 3.7 1 7 8 0.8 1 8 9 0.0 1 9 10 2.0 1 10 11 1.9 2 1 12 0.8 2 2 13 1.1 2 3 14 0.1 2 4 15 -0.1 2 5 16 4.4 2 6 17 5.5 2 7 18 1.6 2 8 19 4.6 2 9 20 3.4 2 10
3.7) Binary ASCII: ASCII, which is short for American Standard Code for Information Interchange is a character-encoding scheme originally based on English alphabet that encodes 128 specified characters – the numbers 0 – 9, the letters a – z and A – Z, some basic punctuation symbols, some control codes that originated with teletype machines and a bland space – into the 7-bit binary integers. It represents text in computers, communications equipment, and other devices that use text. To convert the ASCII code to binary character (part):
| Letter | ASCII Code | Binary | Letter | ASC II Code | Binary |
| a | 097 | 01100001 | A | 065 | 01000001 |
| b | 098 | 01100010 | B | 066 | 01000010 |
| c | 099 | 01100011 | C | 067 | 01000011 |
| d | 100 | 01100100 | D | 068 | 01000100 |
| e | 101 | 01100101 | E | 069 | 01000101 |
| Dec | Hex | Binary |
| 0 | 00 | 00000000 |
| 1 | 01 | 00000001 |
| 2 | 02 | 00000010 |
| 3 | 03 | 00000011 |
| 4 | 04 | 00000100 |
| 5 | 05 | 00000101 |
| 6 | 06 | 00000110 |
| 7 | 07 | 00000111 |
| 8 | 08 | 00001000 |
| 9 | 09 | 00001001 |
| 10 | 0A | 00001010 |
| 11 | 0B | 00001011 |
| 12 | 0C | 00001100 |
| 13 | 0D | 00001101 |
| 14 | 0E | 00001110 |
| 15 | 0F | 00001111 |
| 16 | 10 | 00010000 |
3.8) Handling: the process of ensuring that the research data is stored, archived or disposed off in a safe and secure manner during and after the conclusion of a research project. This includes the development of policies and procedures to manage data handled electronically as well as through non-electronic means.
- Data handling is important in ensuring the integrity of research data since it addresses concerns related to confidentially, security, and preservation/retention of research data. Proper planning for data handling can also result in efficient and economical storage, retrieval, and disposal of data. In the case of data handled electronically, data integrity is a primary concern to ensure that recorded data is not altered, erased, lost or accessed by unauthorized users.
- Data handling issues encompass both electronic as well as non-electronic systems, such as paper files, journals, and laboratory notebooks. Electronic systems include computer workstations and laptops, personal digital assistants (PDA), storage media such as videotape, diskette, CD, DVD, memory cards, and other electronic instrumentation. These systems may be used for storage, archival, sharing, and disposing off data, and therefore, require adequate planning at the start of a research project so that issues related to data integrity can be analyzed and addressed early on.
Issues needed to be considered in ensuring integrity of data handled:
- Type of data handled and its impact on the environment (especially if it is on a toxic media);
- Type of media containing data and its storage capacity, handling and storage requirements, reliability, longevity (in the case of degradable medium), retrieval effectiveness, and ease of upgrade to newer media;
- Data handling responsibilities/privileges, that is, who can handle which portion of data, at what point during the project, for what purpose, etc;
- Data handling procedures that describe how long the data should be kept, and when, how, and who should handle data for storage, sharing, archival, retrieval and disposal purposes.
Applications
4.1) This article (http://en.wikibooks.org/wiki/OpenClinica_User_Manual) presents a comprehensive introduction to EDC (electronic data capture) in clinical research. It contains a series of guides to help users learn how to use OpenClinica for clinical data management. This user manual serves a perfect introduction to OpenClinica and equips researches with background information, specific data management procedures, study construction and system maintenance for data management in clinical studies.
4.2) This article (http://pipeline.loni.usc.edu/learn/user-guide/building-a-workflow/#XNAT) talked about issues regarding building the workflow with LONI Pipeline. It introduced dragging and connecting in the modules and how data is managed with pipeline. It also illustrates usage of pipeline with IDA, NDAR, XNAT and the cloud storage it supported.
4.3) This article (http://onlinepubs.trb.org/onlinepubs/nchrp/cd-22/manual/v2chapter2.pdf) presents a general introduction to data management, analysis tools and analysis mechanics. It illustrated the purpose, steps, considerations and useful tools in data management and introduced the specific steps in data handling. It go through the major steps with examples and illustrated the database handling with consideration of the dataset size. This article offers a comprehensive analysis of data management and would be a good start to implement data management.
4.4) This article (http://www.sciencedirect.com/science/article/pii/S0167819102000947) presents two services that they believe are fundamental to any data grid: reliable, high-speed transport and replica management. Their high-speed transport service, GridFTP, extends the popular FTP protocol with new features required for Data Grid applications, such as striping and partial file access and their replica management service integrates a replica catalog with GridFTP transfers to provide for the creation, registration, location, and management of dataset replicas. The article also presents the design of both services and also preliminary performance results and implementations exploit security and other services provided by the Globus Toolkit.
Software
Data Import/Export in R: http://cran.r-project.org/doc/manuals/r-devel/R-data.html Package ‘rmongodb’ in R (provides an interface to the NoSQL MongoDB database): http://cran.r-project.org/web/packages/rmongodb/ .
Problems
Example 1: Data Streaming:
> library("stream") > dsd <- DSD_Gaussians(k=3, d=3, noise=0.05) > dsd Static Mixture of Gaussians Data Stream With 3 clusters in 3 dimensions > p <- get_points(dsd, n=5) > p
V1 V2 V3
1 0.6930698 0.4082633 0.3857444 2 0.9086645 0.5545718 0.6942871 3 0.6031121 0.5288573 0.5795389 4 0.7590460 0.5485160 0.6484480 5 0.7249682 0.7327056 0.4891291 > p <- get_points(dsd, n=100, assignment=TRUE) > attr(p, "assignment")
[1] 3 3 3 NA 2 2 2 3 1 2 1 2 2 3 1 1 1 3 3 2 3 1 1 2 [25] 3 1 3 3 1 1 3 2 3 3 2 NA 1 1 1 1 1 3 2 2 2 3 1 2 [49] 2 2 2 3 1 NA 1 3 2 2 3 3 3 2 1 2 2 2 1 3 1 1 1 2 [73] 1 3 2 2 3 1 NA 1 2 1 1 3 3 1 1 2 1 3 3 3 2 3 3 2 [97] NA 1 2 1
> plot(dsd, n=500)
Example 2: Import of big data: zips (the data would be available at http://media.mongodb.org/zips.json) import using R/MongoDB: Steps. Install the package of ‘rmongodb’ and import data zips, this data is included in the rmongodb package and can be loaded using the command data(zips):
install.package('rmonogodb') library(rmongodb) data(zips) head(zips)
Output:
> install.packages('rmongodb') trying URL 'http://cran.mtu.edu/bin/macosx/leopard/contrib/2.15/rmongodb_1.6.5.tgz' Content type 'application/x-gzip' length 1291831 bytes (1.2 Mb) opened URL
======================================
downloaded 1.2 Mb
The downloaded binary packages are in /var/folders/k6/3r5dstw5385_b4fzt679pmsr0000gn/T//RtmpWD2wiV/downloaded_packages
> head(zips)
city loc pop state _id
[1,] "ACMAR" Numeric,2 6055 "AL" "35004" [2,] "ADAMSVILLE" Numeric,2 10616 "AL" "35005" [3,] "ADGER" Numeric,2 3205 "AL" "35006" [4,] "KEYSTONE" Numeric,2 14218 "AL" "35007" [5,] "NEW SITE" Numeric,2 19942 "AL" "35010" [6,] "ALPINE" Numeric,2 3062 "AL" "35014"
References
Statistical inference / George Casella, Roger L. Berger http://mirlyn.lib.umich.edu/Record/004199238
Sampling / Steven K. Thompson. http://mirlyn.lib.umich.edu/Record/004232056
Sampling theory and methods / S. Sampath. http://mirlyn.lib.umich.edu/Record/004133572
Data Handling http://ori.hhs.gov/education/products/n_illinois_u/datamanagement/dhtopic.html