Difference between revisions of "SMHS SciVisualization"
| Line 1,018: | Line 1,018: | ||
healthGraph.in.network <- delete.vertices(healthGraph.network, healthGraph.out.network) | healthGraph.in.network <- delete.vertices(healthGraph.network, healthGraph.out.network) | ||
#exclude them from the graph | #exclude them from the graph | ||
| + | |||
| + | # Plot the data by specifying certain details about the graph, e.g., separate some nodes (people) by color: | ||
| + | V(healthGraph.in.network)$\$$color <- ifelse(V(healthGraph.in.network)$\$$name=='CA', 'blue', 'red') | ||
| + | #useful for highlighting certain people. Works by matching the name attribute of the vertex to the one specified in the 'ifelse' expression | ||
| + | # We can also color the connecting edges differently depending on the 'Stage': | ||
| + | E(healthGraph.in.network)$\$$color<-ifelse(E(healthGraph.in.network)$\$$Stage>3, "red", "grey") | ||
| + | # or depending on the different diagnostic agreement labels ('DxAgreement'): | ||
| + | E(healthGraph.in.network)$\$$color<-ifelse(E(healthGraph.in.network)$\$$DxAgreement =='X', "red", ifelse(E(healthGraph.in.network)$\$$DxAgreement=='Y', "blue", "grey")) | ||
| + | |||
| + | |||
| + | # Note: the example uses nested ifelse expressions which can be improved | ||
| + | # Additional attributes like size can be further specified in an analogous manner: | ||
| + | V(healthGraph.in.network)$\$$size<-degree(healthGraph.in.network)/10 | ||
| + | #here the size of the vertices is specified by the degree of the vertex, so that people supervising more have get proportionally bigger dots. Getting the right scale gets some playing around with the parameters of the scale function (from the 'base' package) | ||
| + | |||
| + | # Note that if the same attribute is specified beforehand and inside the function, the former will be overridden. | ||
| + | # And finally the plot itself: | ||
| + | par(mai=c(0,0,1,0)) | ||
| + | #this specifies the size of the margins, default settings leave too much free space on all sides | ||
| + | plot(healthGraph.in.network, #the graph to be plotted | ||
| + | layout=layout.fruchterman.reingold, # the layout method. see the igraph documentation for details | ||
| + | main='Onco Physician Network Example', #specifies the title | ||
| + | vertex.label.dist=0.5, #puts the name labels slightly off the dots | ||
| + | vertex.frame.color='blue', #the color of the border of the dots | ||
| + | vertex.label.color='black', #the color of the name labels | ||
| + | vertex.label.font=2, #the font of the name labels | ||
| + | vertex.label=V(healthGraph.in.network)$\$$name, #specifies the labels of the vertices | ||
| + | vertex.label.cex=1 #specifies the size of the font of the labels | ||
| + | ) | ||
| + | |||
| + | # Save or export the plot as a metafile to the clipboard, a pdf or png (and other formats). | ||
| + | png(filename="org_network.png", height=1900, width=1200) #call the png writer | ||
| + | # alternatively print to high-res PDF file # pdf(file="org_network.pdf") | ||
| + | |||
| + | #run the plot | ||
| + | |||
| + | dev.off() #don’t forget to close the device | ||
| + | |||
| + | ===Pathway analysis=== | ||
| + | |||
| + | Pathway analysis is a technique that reduces complexity and increased explanatory power in studies examining underlying biological structure of differentially expressed genes and proteins. | ||
| + | |||
| + | # install package | ||
| + | # install.packages("dendsort") – contains the “” dataset | ||
| + | # Data: Sample data matrix from the integrated pathway analysis of gastric cancer from the | ||
| + | # Cancer Genome Atlas (TCGA) study. A multivariate table obtained from the integrated pathway analysis | ||
| + | # of gastric cancer from the Cancer Genome Atlas (TCGA) study. Each column represents a pathway | ||
| + | # <u><b>consisting of a set of genes and each row represents a cohort of samples based on specific clinical | ||
| + | # or genetic features.</b></u> For each pair of a pathway and a feature, a continuous value of between | ||
| + | # 1 and -1 is assigned to score positive or negative association, respectively. | ||
| + | # A data frame with <u><b>215</b></u> rows and <u><b>117 variables</b></u> | ||
| + | |||
| + | library("dendsort") | ||
| + | data(sample_tcga) | ||
| + | dataTable <- t(sample_tcga) | ||
| + | head(dataTable) | ||
| + | write.csv(dataTable, "E:\\Ivo.dir\\Research\\UMichigan\\Education_Teaching_Curricula\\2015_2016\\HS_853_Fall_2015\\Modules_docx\\data\\03_TCGA_Data_117x215.csv") | ||
| + | # data.new <- read.csv("https://umich.instructure.com/files/330393/download?download_frd=1") | ||
| + | |||
| + | # install SPIA package: http://bioconductor.org/packages/2.6/bioc/html/SPIA.html | ||
| + | # source("http://bioconductor.org/biocLite.R") | ||
| + | # biocLite("SPIA") | ||
| + | library("SPIA") | ||
| + | |||
| + | # “top” Colorectal cancer dataset provided by SPIA package. | ||
| + | data(Vessels) | ||
| + | head(top) | ||
| + | # pathway analysis based on combined evidence; | ||
| + | # use nB=2000 or more for more accurate results | ||
| + | res<-spia(de=DE_Vessels,all=ALL_Vessels,organism="hsa",nB=500,plots=FALSE,beta=NULL,verbose=FALSE) | ||
| + | #make the output fit this screen | ||
| + | res$\$$Name=substr(res$\$$Name,1,10) | ||
| + | #show first 15 pathways, omitting KEGG links | ||
| + | res[1:15,-12] | ||
| + | |||
| + | <b>GIS/Distortion mapping</b> | ||
| + | |||
| + | # install the R GISTools package | ||
| + | # install.packages("GISTools") | ||
| + | library("GISTools") | ||
| + | data(georgia) | ||
| + | … | ||
| + | |||
| + | Java Applet: http://www.socr.ucla.edu/htmls/SOCR_Cartograhy.html | ||
| + | Activities: http://wiki.stat.ucla.edu/socr/index.php/SOCR_Cartography_Project | ||
| + | |||
| + | |||
| + | <b>Circos Connectogram/Table visualization</b> | ||
| + | |||
| + | Circular chord/ribbon diagrams present a mechanism to visualize numeric tables containing information of directional relations. This type of chart visualizes tables in a circular way. Sectors of the plot is union(rownames(mat), colnames(mat)). When there is no rowname or colname, the chart assigns names for it (rows could be auto-named as "R1", "R2", ... and columns may be named as "C1", "C2"). | ||
| + | |||
| + | • Circos: http://circos.ca | ||
| + | |||
| + | • R circlize: http://cran.r-project.org/web/packages/circlize/circlize.pdf | ||
Revision as of 10:00, 11 April 2016
Contents
Questions
• How and why should we “look” at data?
• What data characteristics are important for exploratory data analytics (EDAs)?
Scientific Data-driven or Simulation-driven visualization methods may be classified in many alternative ways. Visualization techniques can be classified according to many criteria:

• Data Type: structured/unstructured, small/large, complete/incomplete, time/space, ascii/binary, Euclidean/non-Euclidean, etc.
• Task type: Task type is one of the aspects considered in classification of visualization techniques, which provides means of interaction between the researcher, the data and the display software/platform
• Scalability: Visualization techniques are subject to some limitations, such as the amount of data that a particular technique can exhibit
• Dimensionality: Visualization techniques can also be classified according to the number of attributes
• Positioning and Attributes: the distribution of attributes on the chart may affect the interpretation of the display representation, e.g., correlation analysis, where the relative distance among the plotted attributes is relevant for observation
• Investigative Need: the specific scientific question or exploratory interest may also determine the type of visualization:
o Examining the composition of the data
o Exploring the distribution of the data
o Contrasting or comparing several data elements, relations, association
o Unsupervised exploratory data mining
http://www.socr.umich.edu/CSCD/html/Cores/Macore2/SciViz.html
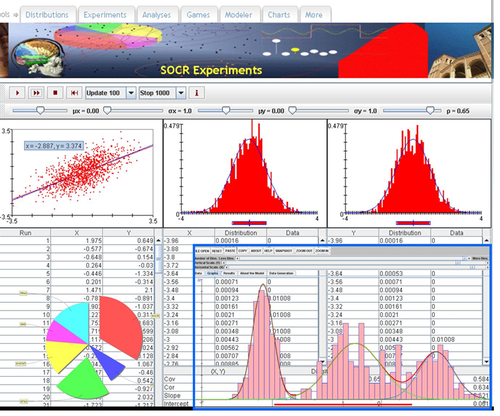
SOCR Charts
• URL: http://socr.umich.edu/html/cha/ (Java applet)
• About/List: http://wiki.stat.ucla.edu/socr/index.php/About_pages_for_SOCR_Chart_List
• Activities: http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials_ChartsActivities
• Data: http://wiki.socr.umich.edu/index.php/SOCR_Data
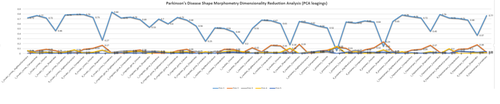
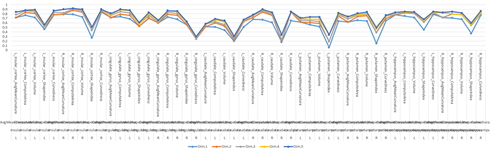
Excel Charts
MS Excel provides a large number of charts that can be used to quickly and effectively render complex multivariate data. For instance, the table below contains the principal component analysis (PCA) of 50 derived neuroimaging biomarkers (region of interest (ROI) by shape morphometry metric). The loadings of these 50 variables on the top 5 (most significant) principal component directions are shown in the table. Experiment with effective visualizations of these data.
| Hemi | ROI | measure | ROI_Measure | Dim.1 | Dim.2 | Dim.3 | Dim.4 | Dim.5 |
|---|---|---|---|---|---|---|---|---|
| L | insular | AvgMeanCurvature | L_insular_cortex_AvgMeanCurvature | 0.72 | 0 | 0.06 | 0.06 | 0 |
| L | insular | ComputeArea | L_insular_cortex_ComputeArea | 0.77 | 0.06 | 0.04 | 0.01 | 0 |
| L | insular | Volume | L_insular_cortex_Volume | 0.72 | 0.09 | 0.04 | 0.03 | 0.01 |
| L | insular | ShapeIndex | L_insular_cortex_ShapeIndex | 0.46 | 0.06 | 0.01 | 0.02 | 0.01 |
| L | insular | Curvedness | L_insular_cortex_Curvedness | 0.78 | 0 | 0.05 | 0.03 | 0.01 |
| R | insular | AvgMeanCurvature | R_insular_cortex_AvgMeanCurvature | 0.79 | 0 | 0.03 | 0.08 | 0 |
| R | insular | ComputeArea | R_insular_cortex_ComputeArea | 0.79 | 0.09 | 0.03 | 0.01 | 0 |
| R | insular | Volume | R_insular_cortex_Volume | 0.73 | 0.11 | 0.03 | 0.03 | 0 |
| R | insular | ShapeIndex | R_insular_cortex_ShapeIndex | 0.27 | 0.17 | 0 | 0.07 | 0 |
| R | insular | Curvedness | R_insular_cortex_Curvedness | 0.84 | 0.02 | 0.03 | 0.01 | 0 |
| L | cingulate | AvgMeanCurvature | L_cingulate_gyrus_AvgMeanCurvature | 0.72 | 0 | 0.05 | 0.02 | 0.02 |
| L | cingulate | ComputeArea | L_cingulate_gyrus_ComputeArea | 0.74 | 0.05 | 0.06 | 0.04 | 0.01 |
| L | cingulate | Volume | L_cingulate_gyrus_Volume | 0.69 | 0.08 | 0.05 | 0.05 | 0.01 |
| L | cingulate | ShapeIndex | L_cingulate_gyrus_ShapeIndex | 0.53 | 0 | 0.05 | 0 | 0.03 |
| L | cingulate | Curvedness | L_cingulate_gyrus_Curvedness | 0.7 | 0.01 | 0.05 | 0.04 | 0.03 |
| R | cingulate | AvgMeanCurvature | R_cingulate_gyrus_AvgMeanCurvature | 0.6 | 0 | 0.02 | 0.03 | 0.01 |
| R | cingulate | ComputeArea | R_cingulate_gyrus_ComputeArea | 0.73 | 0.06 | 0.04 | 0.03 | 0.01 |
| R | cingulate | Volume | R_cingulate_gyrus_Volume | 0.68 | 0.09 | 0.04 | 0.04 | 0.01 |
| R | cingulate | ShapeIndex | R_cingulate_gyrus_ShapeIndex | 0.56 | 0.01 | 0.05 | 0 | 0.01 |
| R | cingulate | Curvedness | R_cingulate_gyrus_Curvedness | 0.25 | 0 | 0.01 | 0.04 | 0 |
| L | caudate | AvgMeanCurvature | L_caudate_AvgMeanCurvature | 0.52 | 0 | 0.05 | 0 | 0.01 |
| L | caudate | ComputeArea | L_caudate_ComputeArea | 0.51 | 0.09 | 0.03 | 0.04 | 0.02 |
| L | caudate | Volume | L_caudate_Volume | 0.44 | 0.09 | 0.03 | 0.06 | 0.03 |
| L | caudate | ShapeIndex | L_caudate_ShapeIndex | 0.2 | 0.03 | 0.04 | 0.04 | 0 |
| L | caudate | Curvedness | L_caudate_Curvedness | 0.51 | 0.12 | 0.02 | 0.01 | 0.01 |
| R | caudate | AvgMeanCurvature | R_caudate_AvgMeanCurvature | 0.68 | 0.04 | 0.04 | 0.02 | 0 |
| R | caudate | ComputeArea | R_caudate_ComputeArea | 0.67 | 0.17 | 0.03 | 0.02 | 0.01 |
| R | caudate | Volume | R_caudate_Volume | 0.61 | 0.16 | 0.02 | 0.03 | 0.01 |
| R | caudate | ShapeIndex | R_caudate_ShapeIndex | 0.18 | 0.02 | 0.03 | 0.11 | 0 |
| R | caudate | Curvedness | R_caudate_Curvedness | 0.65 | 0.19 | 0.01 | 0 | 0 |
| L | putamen | AvgMeanCurvature | L_putamen_AvgMeanCurvature | 0.62 | 0 | 0.04 | 0.03 | 0.02 |
| L | putamen | ComputeArea | L_putamen_ComputeArea | 0.56 | 0.05 | 0.04 | 0.03 | 0.05 |
| L | putamen | Volume | L_putamen_Volume | 0.52 | 0.07 | 0.04 | 0.05 | 0.05 |
| L | putamen | ShapeIndex | L_putamen_ShapeIndex | 0.06 | 0.13 | 0 | 0.15 | 0 |
| L | putamen | Curvedness | L_putamen_Curvedness | 0.64 | 0.11 | 0.03 | 0.01 | 0.03 |
| R | putamen | AvgMeanCurvature | R_putamen_AvgMeanCurvature | 0.62 | 0 | 0.07 | 0.04 | 0.01 |
| R | putamen | ComputeArea | R_putamen_ComputeArea | 0.66 | 0.08 | 0.03 | 0.01 | 0.03 |
| R | putamen | Volume | R_putamen_Volume | 0.64 | 0.12 | 0.03 | 0.02 | 0.03 |
| R | putamen | ShapeIndex | R_putamen_ShapeIndex | 0.15 | 0.24 | 0 | 0.08 | 0.03 |
| R | putamen | Curvedness | R_putamen_Curvedness | 0.65 | 0.05 | 0.05 | 0 | 0.02 |
| L | hippocampus | AvgMeanCurvature | L_hippocampus_AvgMeanCurvature | 0.78 | 0 | 0.01 | 0.04 | 0 |
| L | hippocampus | ComputeArea | L_hippocampus_ComputeArea | 0.75 | 0.07 | 0.01 | 0 | 0.02 |
| L | hippocampus | Volume | L_hippocampus_Volume | 0.72 | 0.09 | 0.01 | 0.01 | 0.01 |
| L | hippocampus | ShapeIndex | L_hippocampus_ShapeIndex | 0.45 | 0.17 | 0 | 0.04 | 0.02 |
| L | hippocampus | Curvedness | L_hippocampus_Curvedness | 0.79 | 0.03 | 0.01 | 0 | 0.02 |
| R | hippocampus | AvgMeanCurvature | R_hippocampus_AvgMeanCurvature | 0.72 | 0 | 0 | 0.1 | 0.01 |
| R | hippocampus | ComputeArea | R_hippocampus_ComputeArea | 0.71 | 0.09 | 0 | 0 | 0.05 |
| R | hippocampus | Volume | R_hippocampus_Volume | 0.68 | 0.1 | 0 | 0 | 0.04 |
| R | hippocampus | ShapeIndex | R_hippocampus_ShapeIndex | 0.37 | 0.18 | 0 | 0.02 | 0.03 |
| R | hippocampus | Curvedness | R_hippocampus_Curvedness | 0.77 | 0.03 | 0 | 0.02 | 0.04 |

R-Charts
There are 100’s of packages and 1,000 of different charts, plots and graphs that can be generated using R. Such interactive visualizations enable deeper exploration of data, models and results. JavaScript libraries, e.g., D3, provide advantages for data visualization as these involve HTML5 and are easily shareable online. The R community is developing R interfaces to some popular JavaScript libraries to allow users to create interactive visualizations without detailed knowledge of JavaScript.
Examples of powerful R interactive visualization packages
• ggplot2 – http://ggplot2.org
• ggvis – interactive plots extending the static ggplot2 charts, http://ggvis.rstudio.com
• rCharts – R interface to multiple JavaScript charting libraries, http://rcharts.io
• plotly – transforming ggplot2 charts into interactive plots, https://plot.ly/r
• googleVis – Google Charts using R, http://cran.r-project.org/web/packages/googleVis/vignettes/googleVis_examples.html
• HTMLWidgets
o leaflet – library for creating dynamic maps, supports panning and zooming, annotations, markers, polygons, etc. http://www.htmlwidgets.org/showcase_leaflet.html
o dygraphs – provides mechanism for charting time-series data, supports interactive navigation features including series/point highlighting, zooming, and panning, http://www.htmlwidgets.org/showcase_dygraphs.html
o networkD3 – library for creating D3 network graphs including force directed networks, Sankey diagrams, and Reingold-Tilford tree networks, http://www.htmlwidgets.org/showcase_networkD3.html
o DataTables – displays R matrices or data frames as interactive HTML tables that support filtering, pagination, and sorting, http://www.htmlwidgets.org/showcase_datatables.html
o Rthreejs – features 3D scatterplots and globes based on WebGL, http://www.htmlwidgets.org/showcase_threejs.html
• Other R graphic examples
o To write out plots out to file use:
# pdf() command all graphs are redirected to test.pdf. Also works with other common formats: jpeg, png, ps, tiff.
pdf("C:\\Users\\Dinov\\Desktop\\test.pdf"); plot(1:100, 1:100); dev.off()
# Generates Scalable Vector Graphics (SVG) that can be edited by vector graphics software
svg("test.svg"); plot(1:100, 1:100); dev.off()
Paired ScatterPlots
set.seed(100)
x <- matrix(runif(50), ncol=5, dimnames=list(letters[1:10], LETTERS[1:5]))
describe(x) # library("Hmisc")
plot(x[,1], x[,2], pch=20, col="red", main="Symbols and Labels")
text(x[,1]+0.03, x[,2], rownames(x))
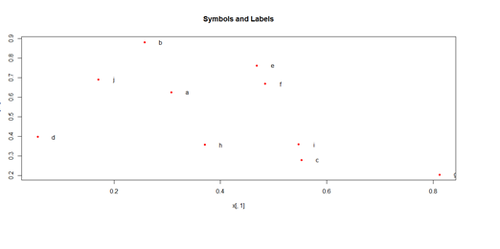
pairs(x)
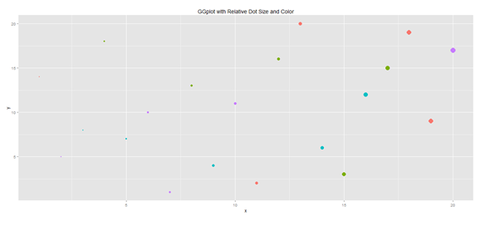
Another way to generate scatterplots is by using ggplot:
# library(ggplot2)
x <- sample(1:20, 20); y <- sample(1:20, 20); cat <- rep(c("A", "B", "C", "D"), 5)
#vs. cat <- rep(c("A", "B", "C", "D"), each=5)
plot.1 <- qplot(x, y, geom="point", size=5*x, color=cat, main="GGplot with Relative Dot Size and Color") + theme(legend.position = "topleft")
print(plot.1)
# Use Case-Studies: https://umich.instructure.com/courses/38100/files/folder/Case_Studies # Case_03_MentalHealthServicesSurvey # data1 <- read.table('https://umich.instructure.com/files/399128/download?download_frd=1&verifier=AG2e9QUKUm1jvDBpkX7D9jbEjKNc4irA0ECk0f7p', header=T) head(data1) attach(data1) # library("Hmisc") describe(data1)
plot(data1[,3], data1[,4], pch=20, col="red", main="Symbols and Labels") # text(data1 [,3]+0.03, data1 [,4], rownames(data1)) plot.1 <- qplot(x, y, geom="point", size=5*x, color=cat, main="GGplot with Relative Dot Size and Color") + theme(legend.position = "topleft") print(plot.1)
# redo plots using majorfundtype FacilityType Ownership Focus # pairs(data1, na.action=na.omit)
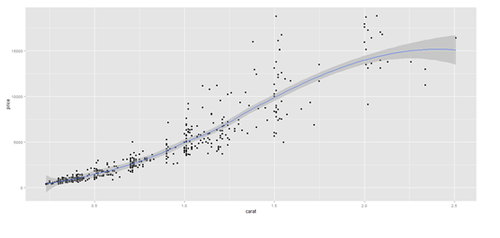
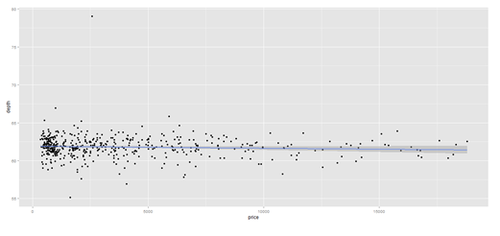
# Scatterplot with regression line. Use the “diamonds” dataset, which is a data frame with # 53,940 rows and 10 variables () # describe(diamonds)
# Use Case-Studies: https://umich.instructure.com/courses/38100/files/folder/Case_Studies # CaseStudy01_Divorce_YoungAdults # data1 <- read.csv('https://umich.instructure.com/files/399118/download?download_frd=1&verifier=ESACv31KcyiHbkPZPuT8Oo4V7XzPtgTTbs6PQLTv', header=T) attach(data1) # plot variables: DIVYEAR momint dadint momclose depression livewithmom gethitched
set.seed(110)
# par(mfrow=c(1,2))
data.2 <- diamonds[sample(nrow(diamonds), 500), ]
plot.2 <- qplot(price, depth, data = data.2, geom = c("point", "smooth"), method = "lm")
plot.3 <- qplot(carat, price, data=data.2, geom=c("point", "smooth"), span=0.4)
print(plot.2); print(plot.3)
Barplots
x <- matrix(runif(50), ncol=5, dimnames=list(letters[1:10], LETTERS[1:5]))
barplot(x[1:4,], ylim=c(0, max(x[1:4,])+0.3), beside=TRUE, legend.text = letters[1:4],
args.legend = list(x = "topleft"))
text(labels=round(as.vector(as.matrix(x[1:4,])),2), x=seq(1.5, 21, by=1) + sort(rep(c(0,1,2,3,4), 4)), y=as.vector(as.matrix(x[1:4,]))+0.1)
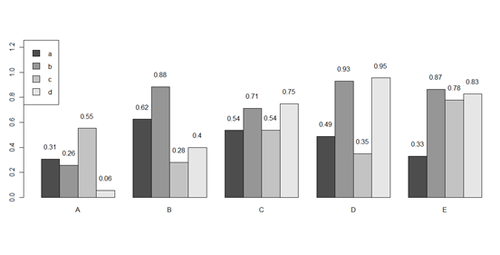
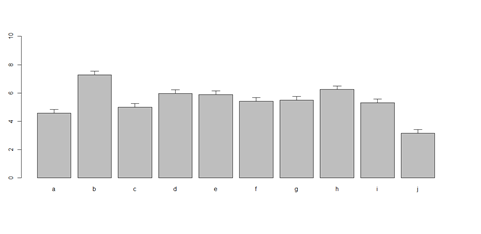
# to put error bars on barplot:
# 10 rows (a, b, c, …): bar <- barplot(m <- rowMeans(x) * 10, ylim=c(0, 10)) stdev <- sd(t(x)) arrows(bar, m, bar, m + stdev, length=0.15, angle = 90)
# Case_04_ChildTrauma
# data1 <- read.table('https://umich.instructure.com/files/399129/download?download_frd=1&verifier=Hmv0YW2Kie5ZTV9CKBUNArSHR66f3GWSmVzZDBxc', header=T)
attach(data1)
head(x)
head(data1)
# plot data
data2 <- data1[,-5] # remove the 5th columns text
data1 <- data2[,-5] # remove the 6th columns text
# or data1 <- data1[,c(-5,-6)]
data2 <- as.data.frame(data1) Blacks <- data2[which(data2$\$$race=="black"),] Other <- data2[which(data2$\$$race=="other"),] Hispanic <- data2[which(data2$\$$race=="hispanic"),] White <- data2[which(data2$\$$race=="white"),]
A <- c(mean(Blacks$\$$age), mean(Blacks$\$$service))
#colnames(A) <- c("age "," service ")
B <- c(mean(Other$\$$age), mean(Other$\$$service))
C <- c(mean(Hispanic$\$$age), mean(Hispanic$\$$service))
D <- c(mean(White$\$$age), mean(White$\$$service))
x <- cbind(A, B, C, D)
bar <- barplot(x[1:2,], ylim=c(0, max(x[1:2,])+2.0), beside=TRUE,
legend.text = c("age","service") , args.legend = list(x = "right"))
text(labels=round(as.vector(as.matrix(x[1:2,])),2), x=seq(1.4, 21, by=1.5), #y=as.vector(as.matrix(x[1:2,]))+0.3)
y=11.5)
m <- x; stdev <- sd(t(x)) arrows(bar, m, bar, m + stdev, length=0.15, angle = 90)
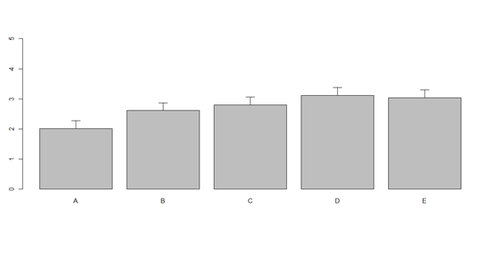
barplot(as.matrix(data1[1:4,]), ylim=c(0, max(data1[1:4,])+0.3), beside=TRUE, legend.text = data1[1:4,1], args.legend = list(x = "topleft")) text(labels=round(as.vector(as.matrix(data1[1:4,])),2), x=seq(1.5, 21, by=1), y=as.vector(as.matrix(data1[1:4,]))+0.1)
# Columns (A, B, C, D, E): bar <- barplot(m <- colMeans(x) * 5, ylim=c(0, 5)) stdev <- sd(t(x)) arrows(bar, m, bar, m + stdev, length=0.15, angle = 90)
Histograms and Density Plots
hist(x, freq=TRUE, breaks=10)
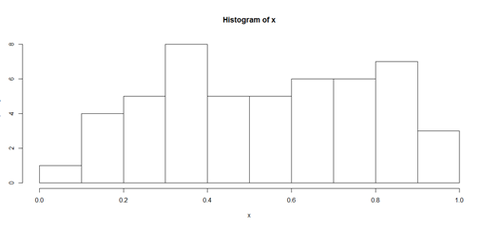
plot(density(x), lwd = 10, col="green")
Pie Chart
# first , “A”, and second, “B”, columns par (mfrow=c(1,2)) pie(x[,1], col=rainbow(length(x[,1]), start=0.1, end=0.8), clockwise=TRUE)
pie(x[,1], col=rainbow(length(x[,1]), start=0.1, end=0.8), clockwise=TRUE)
pie(x[,2], col=rainbow(length(x[,2]), start=0.1, end=0.8), clockwise=TRUE)
legend("topleft", legend=row.names(x), cex=1.3, bty="n", pch=15, pt.cex=1.8, col=rainbow(length(x[,2]), start=0.1, end=0.8), ncol=1)
You can export the data: write.table(x, " ", "data.txt") # copy-paste it in SOCR Pie chart to generate another Pie view of data
Line Plots Using ggplot
head(diamonds)
| Carat | Cut | Color | Clarity | Depth | Table | Price | X | Y | Z | |
| 1 | 0.23 | Ideal | E | SI2 | 61.5 | 55 | 326 | 3.95 | 3.98 | 2.43 |
| 2 | 0.21 | Premium | E | SI1 | 59.8 | 61 | 326 | 3.89 | 3.84 | 2.31 |
| 3 | 0.23 | Good | E | VS1 | 56.9 | 65 | 237 | 4.05 | 4.07 | 2.31 |
| 4 | 0.29 | Premium | I | VS2 | 62.4 | 58 | 334 | 4.2 | 4.23 | 2.63 |
| 5 | 0.31 | Good | J | SI2 | 63.3 | 58 | 335 | 4.34 | 4.35 | 4.75 |
| 6 | 0.24 | VeryGood | J | VVS2 | 62.8 | 57 | 336 | 3.94 | 3.96 | 2.48 |
plot.2 <- ggplot(diamonds, aes(carat, price, group=cut, color=cut)) + geom_line() print(plot.2)
plot.2 <- ggplot(data1, aes(age, service, group=race, color=race)) + geom_line() print(plot.2)
# Faceting plot (geometrically, faceting (or facetting) is the process of removing parts of a polygon, polyhedron or polytope, without creating any new vertices) plot.3 <- ggplot(diamonds, aes(carat, price)) + geom_line(aes(color=cut), size=1) + facet_wrap(~cut, ncol=1) print(plot.3)
Barplots with ggplot
plot.4 <- ggplot(diamonds, aes(cut, fill=cut)) + geom_bar() + facet_grid(. ~ clarity) print(plot.4)
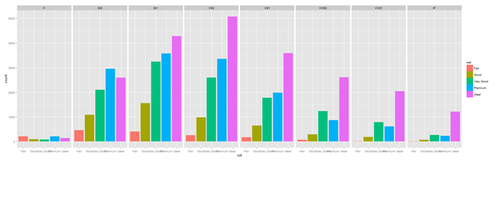
New_var <- service+rnorm(1000, 0,1)
data1$\$$New_var <- int(New_var)
plot.4 <- ggplot(data1, aes(race, fill= traumatype)) + geom_bar() + facet_grid(. ~ New_var)
print(plot.4)
plot.4a <- ggplot(diamonds, aes(color, price/carat, fill=color)) + geom_boxplot()
print(plot.4a)
<center>[[Image:SMHS_SciVisualization20.png|500px]] </center>
==='"`UNIQ--h-9--QINU`"'Jitter plot===
plot.5 <- ggplot(diamonds, aes(color, price/carat)) + geom_jitter(alpha = I(1 / 2), aes(color=color))
print(plot.5)
<center>[[Image:SMHS_SciVisualization21.png|500px]] </center>
==='"`UNIQ--h-10--QINU`"'Density Plots===
plot.6 <- ggplot(diamonds, aes(carat, size=2)) + geom_density(aes(color = cut))
print(plot.6)
<center>[[Image:SMHS_SciVisualization22.png|500px]] </center>
plot.6 <- ggplot(data1, aes(age, size=2)) + geom_density(aes(color = traumatype))
print(plot.6)
plot.7 <- ggplot(diamonds, aes(carat, size=2)) + geom_density(aes(fill = color))
print(plot.7)
<center>[[Image:SMHS_SciVisualization23.png|500px]] </center>
plot.8 <- ggplot(diamonds, aes(x=carat, size=1)) + geom_histogram(aes(y = price), binwidth=0.2) + geom_density()
print(plot.8)
<center>[[Image:SMHS_SciVisualization24.png|500px]] </center>
plot.8a <- ggplot(diamonds, aes(x=carat, size=1)) + geom_histogram(aes(y = price), stat="identity") + geom_density()
print(plot.8a)
<center>[[Image:SMHS_SciVisualization25.png|500px]] </center>
==='"`UNIQ--h-11--QINU`"'Heatmaps===
# Generating Dendogram Association Heatmap Plot (Genotype vs. Imaging phenotype
# http://stat.ethz.ch/R-manual/R-patched/library/stats/html/heatmap.html
# http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4005931/
AD_Associations_Data <- read.table("https://umich.instructure.com/files/330387/download?download_frd=1&verifier=gLk2ADgrLhXGeknI6mqIeJugi2ODr8RARsQlBUMe", header=TRUE, row.names=1, sep=",", dec=".")
MCI_Associations_Data <- read.table("https://umich.instructure.com/files/330390/download?download_frd=1&verifier=FczlJD6ISRPZhu69xvHuoZHx2c7gXX9YEvvPCTBG", header=TRUE, row.names=1, sep=",", dec=".")
NC_Associations_Data <- read.table("https://umich.instructure.com/files/330391/download?download_frd=1&verifier=i2BEtSpmpbrzQUPoA2ST06IzzcaenyVEHRepHSF3", header=TRUE, row.names=1, sep=",", dec=".")
require(graphics)
require(grDevices)
library(gplots)
AD_Data <- AD_Associations_Data
MCI_Data <- MCI_Associations_Data
NC_Data <- NC_Associations_Data
AD_mat <- as.matrix(AD_Data); class(AD_mat) <- "numeric"
MCI_mat <- as.matrix(MCI_Data); class(MCI_mat) <- "numeric"
NC_mat <- as.matrix(NC_Data); class(NC_mat) <- "numeric"
# set up the rol (rc) and column (cc) colors for each cohort
rcAD <- rainbow(nrow(AD_mat), start = 0, end = 1.0); ccAD<-rainbow(ncol(AD_mat), start = 0, end = 1.0)
rcMCI <- rainbow(nrow(MCI_mat), start = 0, end=1.0); ccMCI<-rainbow(ncol(MCI_mat),start=0,end=1.0)
rcNC <- rainbow(nrow(NC_mat), start = 0, end = 1.0); ccNC<-rainbow(ncol(NC_mat), start = 0, end = 1.0)
# set up 1x3 graph display - par (mfrow=c(1,3)) – does not work with ‘heatmap’
hvAD <- heatmap(AD_mat, col = cm.colors(256), scale = "column", RowSideColors = rcAD, ColSideColors = ccAD, margins = c(2,2), main="AD Cohort SNP-ROI_volume Association (p_values)")
hvMCI <- heatmap(MCI_mat, col = cm.colors(256), scale = "column", RowSideColors = rcMCI, ColSideColors = ccMCI, margins = c(2,2), main="MCI Cohort SNP-ROI_volume Association (p_values)")
hvNC <- heatmap(NC_mat, col = cm.colors(256), scale = "column", RowSideColors = rcNC, ColSideColors = ccNC, margins = c(2,2), main="NC Cohort SNP-ROI_volume Association (p_values)")
<center>[[Image:SMHS_SciVisualization26.png|500px]] </center>
# Alternatively, we can use the R package gplots
install.packages("gplots")
library(gplots)
## col dendrogram plotted and col reordering done.
# heatmap.2(AD_mat, keysize=2)
## A more decorative heatmap, with z-score scaling along columns
heatmap.2(AD_mat, col=cm.colors(255), scale="column", RowSideColors=rcAD, ColSideColors=ccAD, margin=c(8, 7), xlab="Imaging Biomarkers (ROI volume)", ylab= "Genetics Biomarkers (SNPs)", main="AD Associations Heatmap (SNP-Imaging)", tracecol="green", density="density")
<center>[[Image:SMHS_SciVisualization27.png|500px]] </center>
==='"`UNIQ--h-12--QINU`"'Correlation Plots===
The <b>corrplot</b> package is a graphical display of a correlation matrix, and confidence intervals, with some tools for matrix reordering. There are seven visualization methods (parameter method) in corrplot package, named "circle", "square", "ellipse", "number", "shade", "color", "pie".
# install.packages("corrplot")
library(corrplot)
NC_Associations_Data <- read.table("https://umich.instructure.com/files/330391/download?download_frd=1&verifier=i2BEtSpmpbrzQUPoA2ST06IzzcaenyVEHRepHSF3", header=TRUE, row.names=1, sep=",", dec=".")
M <- cor(NC_Associations_Data)
<center>[[Image:SMHS_SciVisualization28.png|500px]] </center>
==='"`UNIQ--h-13--QINU`"'Hyperbolic Visualization===
• URL: http://socr.umich.edu/html/Navigators.html
• Tools:
<blockquote>o Java/Jar applet: http://socr.umich.edu/html/navigators/HW/jars/SOCR_HW_Viewer.jar</blockquote>
<blockquote>o JavaScript: http://socr.umich.edu/html/navigators/D3/SOCR_D3_Viewer.html (JSON)</blockquote>
• Data Format
<blockquote>o XML data: http://socr.umich.edu/html/navigators/HW/SOCR_HyperTree.xml</blockquote>
<blockquote>o JSON data: http://socr.umich.edu/html/navigators/D3/xml/SOCR_HyperTree.json</blockquote>
• D3 Visualization
<blockquote>o E:\Ivo.dir\Research\UMichigan\Education_Teaching_Curricula\2015_2016\HS_853_Fall_2015\Modules_docx\Tools\TreeViewer_JS</blockquote>
<blockquote> treeJS.json</blockquote>
<blockquote> flareD3.json</blockquote>
<center>[[Image:SMHS_SciVisualization29.png|500px]] </center>
<center>[[Image:SMHS_SciVisualization30.png|500px]] </center>
• URL: https://github.com/mbostock/d3/wiki/Gallery
• Source code: https://github.com/mbostock/d3
<center>[[Image:SMHS_SciVisualization31.png|500px]] </center>
==='"`UNIQ--h-14--QINU`"'Motion Charts===
• Video: http://www.socr.ucla.edu/SOCR_MotionCharts/SOCR_HTML5_MotionChart_Video2.gif
• Java: http://www.socr.ucla.edu/SOCR_MotionCharts/
• HTML5: http://socr.umich.edu/HTML5/MotionChart/
• Activities: http://wiki.socr.umich.edu/index.php/SOCR_MotionCharts
<center>[[Image:SMHS_SciVisualization32.png|500px]] </center>
==='"`UNIQ--h-15--QINU`"'1D/2D/3D signal/area/volume/surface/model/atlas visualization===
• 1D: (See R/SOCR Visualization tools above)
• 2D: http://imagej.nih.gov/ij/
• 3D: http://socr.umich.edu/HTML5/BrainViewer/
<b>Supported File Formats:</b>
Volumes (.nii / .nii.gz / .img&.hdr / .mgh / .mgz / .nrrd)
Shapes (.dx / .vtk / .stl / FreeSurfer)
Fibers (.trk)
<center>[[Image:SMHS_SciVisualization33.png|500px]] </center>
==='"`UNIQ--h-16--QINU`"'Trees and Graphs===
• Trees/Hierarchies and general Graphs
# Install and load the APE package, needed for the phylogenetic tree rendering (as.phylo)
# install.packages("ape")
library("ape")
Load data
# Data: 02_Nof1_Data.csv
data.1 <- read.table("https://umich.instructure.com/files/330385/download?download_frd=1&verifier=DwJUGSd6t24dvK7uYmzA2aDyzlmsohyaK6P7jK0Q ", sep=",", header = TRUE)
head(data.1)
<center>
{| class="wikitable" style="text-align:center; width:35%" border="1"
|-
mydata1
|-
|||ID||Day||Tx||SelfEff||SelfEff25||WPSS||SocSuppt||PMss||PMss3||PhyAct
|-
|1||1||1||1||33||8||0.97||5.00||4.03||1.03||53
|-
|2||1||2||1||33||8||-0.17||3.87||4.03||1.03||73
|-
|3||1||3||0||33||8||0.81||4.84||4.03||1.03||23
|-
|4||1||4||0||33||8||-0.41||3.62||4.03||1.03||36
|-
|5||1||5||1||33||8||0.59||4.62||4.03||1.03||21
|-
|6||1||6||1||33||8||-1.16||2.87||4.03||1.03||0
|}
</center>
Clustering
hc = hclust(dist(data.1), 'ave')
# the agglomeration method can be specified "ward.D", "ward.D2", "single","complete", "average" (= UPGMA), "mcquitty" (= WPGMA),"median" (= WPGMC) or "centroid" (= UPGMC)
# (3) Plot clustering diagram
par (mfrow=c(1,1))
# very simple dendrogram
plot(hc)
<center>[[Image:SMHS_SciVisualization34.png|500px]] </center>
require(graphics)
(x <- identify(hc)) ## Terminate with 2nd mouse button !!
identify(hc, <mark>function(k)</mark> print(table(data.1[k,5])))
You can now cut the tree into branches. You can split the tree into 2 groups, by setting the number of cuts with the k=2 parameter, or by specifying height to cut the tree at (?cutree):
k an integer scalar or vector with the desired number of groups
h numeric scalar or vector with heights where the tree should be cut
cutree(hc, k = 2)
# alternatively specify the height, which is, the value of the criterion associated with the clustering method
# for the particular agglomeration.
cutree(hc, h= 50) # cut at h=50
table(cutree(hc, h= 50)) # cluster distribution
# To identify the number of cases for varying number of clusters we can combine calls to cutree and table
# in a call to <b>sapply</b> -- to see the sizes of the clusters for 2≤ k≤10 cluster-solutions:
# numbClusters=5;
myClusters = sapply(2:10,function(numbClusters)table(cutree(hc, numbClusters)))
names(myClusters) <- paste("Number of Clusters=", 2:10, sep = "")
myClusters
#To see which SubjectIDs are in which clusters:
groups.10 <- cutree(hc, k = 10)
sapply(unique(groups.10),function(g)data.1$\$$ID[groups.10 == g])
#To see which Treatments (Tx) are in which clusters:
groups.2 <- cutree(hc, k = 2)
sapply(unique(groups.2),function(g)data.1$\$$Tx[groups.2 == g])
# drill down deeper
table(groups.2, data.1$\$$Tx)
# For a small number of observations, we can often interpret the cluster solution directly by looking
# at the labels of the observations that are in each cluster.
# This is hard for larger data sets. To characterize clusters we can look at cluster summary statistics,
# like the median, of the variables that were used to perform the cluster analysis broken down
# by the groups that the cluster analysis identified.
The aggregate function will compute stats (e.g., median) on many variables simultaneously.
To look at the median values for the variables we've used in the cluster analysis, broken up by the cluster groups:
aggregate(data.1, list(groups.10),median) # may have to shrink data.1 prior to clustering! # data.2 <- data.1[,-c(1,3)] # Remove ID and Tx variables? aggregate(data.2, list(groups.2),median) # for only 2 clusters
| Group | ID | Day | Tx | SelfEff | SelfEff25 | WPSS | SocSuppt | PMss | PMss3 | PhyAct | |
| 1 | 1 | 14 | 16 | 0 | 20 | -5 | -0.040 | 2.995 | 3.275 | 0.275 | 41 |
| 2 | 2 | 16 | 15 | 1 | 25 | 0 | 0.025 | 3.280 | 3.360 | 0.360 | 104 |
table(groups.2, data.1$\$$<u><b>PhyAct</b></u>)
# library("Hmisc")
describe(data.1$\$$PhyAct)
# It’s useful to add the numbers of observations in each group (aggregate returns a data frame, # that can be manipulated)
df.2 <- aggregate(data.1, list(groups.2),median)
data.frame(Cluster= df.2[,1], Freq=as.vector(table(groups.2)), df.2[,-1])
| Cluster | Freq | ID | Day | Tx | SelfEff | SelfEff25 | WPSS | SocSuppt | PMss | PMss3 | PhyAct | |
| 1 | 1 | 570 | 14 | 16 | 0 | 20 | -5 | -0.040 | 2.995 | 3.275 | 0.275 | 41 |
| 2 | 2 | 330 | 16 | 15 | 1 | 25 | 0 | 0.025 | 3.280 | 3.360 | 0.360 | 104 |
Publications
• This paper examines nasal and bronchial tissue cultures as appropriate in vitro models for the assessment of smoking-induced adverse effects in the respiratory system (doi: 10.1177/1091581814551647), using “hclust” package. No data.
• This paper classified subtypes of gastric cancer based on epidemiologic and histologic and gene expression data. These new classifications of gastric cancer have implications for improving our understanding of disease biology and identification of unique molecular drivers for each gastric cancer subtype (doi: 10.1158/1078-0432.CCR-10-2203).
Repeat the clustering
# using centroids and squared Euclidean distance
# cut the tree into 10 clusters and reconstruct the upper part of the tree from the cluster centers.
hc <- hclust(dist(data.1), "cen")
mem <- cutree(hc, k = 10)
cent <- NULL
for(k in 1:10){
cent <- rbind(cent, colMeans(data.1[mem == k, , drop = FALSE]))
}
hc1 <- hclust(dist(cent), method = "cen", members = table(mem))
opar <- par(mfrow = c(1, 2)) plot(hc, labels = FALSE, hang = -1, main = "Original Tree") plot(hc1, hang = -1, main = "Re-start from 10 clusters") par(opar)
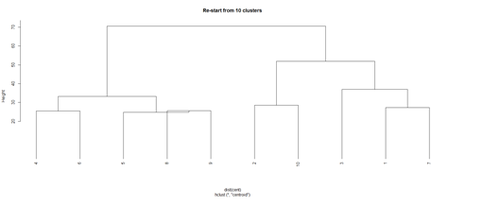
Identify subjects within each of the 10 classes
rect.hclust(hc, h=10)
# To save the cluster numbers to a new variable in the dataset, use the cutree function. # data.1$\$$clusterID <- cutree(hc, 10) data.1$\$$clusterID <- cutree(hc, 10) head(data.1)
| ID | Day | Tx | SelfEff | SelfEff25 | WPSS | SocSuppt | PMss | PMss3 | PhyAct | CluserID | |
| 1 | 1 | 1 | 1 | 33 | 8 | 0.97 | 5.00 | 4.03 | 1.03 | 53 | 1 |
| 2 | 1 | 2 | 1 | 33 | 8 | -0.17 | 3.87 | 4.03 | 1.03 | 73 | 1 |
| 3 | 1 | 3 | 0 | 33 | 8 | 0.81 | 4.84 | 4.03 | 1.03 | 23 | 2 |
| 4 | 1 | 4 | 0 | 33 | 8 | -0.41 | 3.62 | 4.03 | 1.03 | 36 | 2 |
| 5 | 1 | 5 | 1 | 33 | 8 | 0.59 | 4.62 | 4.03 | 1.03 | 21 | 2 |
| 6 | 1 | 6 | 1 | 33 | 8 | -1.16 | 2.87 | 4.03 | 1.03 | 0 | 2 |
Phylogenetic tree diagram
# library("ape")
plot(as.phylo(hc1), use.edge.length = TRUE, type = "fan")
plot(as.phylo(hc), use.edge.length = TRUE, type = "fan", tip.color = hsv(runif(15, 0.65, 0.95), 1, 1, 0.7), label.offset = 1, cex = log(data.1$\$$ID, 10), col = "red")
<center>[[Image:SMHS_SciVisualization36.png|500px]] </center>
<center>[[Image:SMHS_SciVisualization37.png|500px]] </center>
<b>Hands-on Activity (Health Behavior Risks)</b>
# load data CaseStudy09_HealthBehaviorRisks_Data
data.2 <- read.csv("https://umich.instructure.com/files/399182/download?download_frd=1 ", sep=",", header = TRUE)
# Classify the cases using these variables: "AGE_G" "SEX" "RACEGR3" "IMPEDUC" "IMPMRTL"
# "EMPLOY1" "INCOMG" "CVDINFR4" "CVDCRHD4" "CVDSTRK3" "DIABETE3" "RFSMOK3"
# "FRTLT1" "VEGLT1"
data.raw <- data.2[,-c(1,14,17)]
# Does the classification match either of these:
# TOTINDA (Leisure time physical activities per month, 1=Yes, 2=No, 9=Don’t know/Refused/Missing)
# RFDRHV4 (Heavy alcohol consumption, 1=No, 2=Yes, 9=Don’t know/Refused/Missing)
hc = hclust(dist(data.raw), 'ave')
# the agglomeration method can be specified "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC)
# (3) Plot clustering diagram
par (mfrow=c(1,1))
# very simple dendrogram
plot(hc)
summary(data.2$\$$TOTINDA); summary(data.2$\$$RFDRHV4)
cutree(hc, k = 2)
# alternatively specify the height, which is, the value of the criterion associated with the
# clustering method for the particular agglomeration -- cutree(hc, h= 10)
table(cutree(hc, h= 10)) # cluster distribution
# To identify the number of cases for varying number of clusters we can combine calls to cutree and table
# in a call to sapply -- to see the sizes of the clusters for 2≤ k≤10 cluster-solutions:
# numbClusters=4;
myClusters = sapply(2:5,function(numbClusters)table(cutree(hc, numbClusters)))
names(myClusters) <- paste("Number of Clusters=", 2:5, sep = "")
myClusters
#To see which SubjectIDs are in which clusters:
table(cutree(hc, k=2))
groups.k.2 <- cutree(hc, k = 2)
sapply(unique(groups.k.2),function(g)data.2$\$$ID[groups.k.2 == g])
#To see which TOTINDA (Leisure time physical activities per month, 1=Yes, 2=No, 9=Don’t # know/Refused/Missing) & whch RFDRHV4 are in which clusters: groups.k.3 <- cutree(hc, k = 3) sapply(unique(groups.k.3),function(g)data.2$\$$TOTINDA [groups.k.3 == g]) sapply(unique(groups.k.3),function(g)data.2$\$$RFDRHV4[groups.k.3 == g])
# Perhaps there are intrinsically 3 groups here e.g., 1, 2 and 9 … groups.k.3 <- cutree(hc, k = 3) sapply(unique(groups.k.3),function(g)data.2$\$$TOTINDA [groups.k.3 == g]) sapply(unique(groups.k.3),function(g)data.2$\$$RFDRHV4 [groups.k.3 == g])
# Note that there is quite a dependence between the outcome variables … plot(data.2$\$$RFDRHV4, data.2$\$$TOTINDA)
# drill down deeper
table(groups.k.3, data.2$\$$RFDRHV4)
# To characterize clusters we can look at cluster summary statistics,
# like the median, of the variables that were used to perform the cluster analysis broken down
# by the groups that the cluster analysis identified. The aggregate function will compute stats
# (e.g., median) on many variables simultaneously. To look at the median values for the variables
# we've used in the cluster analysis, broken up by the cluster groups:
aggregate(data.2, list(groups.k.3),median)
==='"`UNIQ--h-18--QINU`"'Complex Network Visualization===
# Install package
# install.packages("igraph")
library("igraph")
# build a simple graph
g <- graph( c(1,2, 1,3, 2,3, 3,4), n=10)
plot(g)
summary(g); g; is.igraph(g); is.directed(g); vcount(g); ecount(g)
<center>[[Image:SMHS_SciVisualization38.png|500px]] </center>
<center>[[Image:SMHS_SciVisualization39.png|500px]] </center>
plot(g, layout=layout.circle)
plot(g, layout=layout.fruchterman.reingold)
plot(g, layout=layout.graphopt)
plot(g, layout=layout.kamada.kawai, vertex.color="cyan")
# Interactive
tkplot(g, layout=layout.kamada.kawai)
# 3D plot
rglplot(g, layout=layout.kamada.kawai(g))
# Dataset 1: Coappearance network in the novel “les miserablese”.
# D. E. Knuth, The Stanford GraphBase: A Platform for Combinatorial Computing, Addison-Wesley, Reading,
# MA (1993).
# The data contains the weighted network of coappearances of characters in Victor Hugo's novel "Les
# Miserables". Nodes represent characters as indicated by the labels and edges connect any pair of
# characters that appear in the same chapter of the book. The values on the edges are the number of
# such coappearances.
# Alternatively, we can use a directed, weighted network representing the neural network of the nematode
# C. Elegans. D. Watts and S. Strogatz, Nature 393, 440-442 (1998). The file celegansneural.gml describes a
# weighted, directed network where the nodes have been renumbered to be consecutive.
# Edge weights are the weights given by Watts.
install.packages("rgl")
library("igraph")
g<-read.graph("C:\\Users\\Dinov\\Desktop\\celegansneural.gml",format=c("gml"))
g
plot(g, layout=layout.graphopt)
data_g <- read.table("https://umich.instructure.com/files/330389/download?download_frd=1&verifier=u1jqCGS8AAU0MsO5ffLCyvVFYXXAflpdLtg8RXhk", sep=" ", header = FALSE)
data_g_mat <- as.matrix(data_g, byrow=TRUE, nc=2)
g_miserab <- graph.edgelist(data_g_mat, dir=FALSE)
summary(g_miserab)
plot(g_miserab, layout=layout.graphopt)
# rglplot(g_miserab, layout=layout.kamada.kawai(g_miserab))
# to name the vertices and and plot the graph of the first 10 vertices
V(g_miserab)$name
g_miserab.1 <- graph.ring(10)
V(g_miserab.1)$name <- sample(letters, vcount(g_miserab.1))
plot(g_miserab.1, layout=layout.graphopt)
# compute the node adjacency matrix
g <- g_miserab; as_adjacency_matrix(g)
E(g)$weight <- runif(ecount(g))
W <- get.adjacency(g, attr="weight")
W
<b>Social Network Analysis Example</b>
# free memory
# rm(list = ls())
# gc()
# load termDocMatrix dataset
# These data include Twitter text data of @RDataMining representing a general social network analysis
# example. The terms represent people and the tweets represent LinkedIn groups.
# The term-document matrix can be viewed as the group membership of people.
# We may want to build a network of terms based on their co-occurrence in the same tweets,
# similarly to a network of people based on their group membership.
# https://umich.instructure.com/files/541336/download?download_frd=1
load("E:\\Ivo.dir\\Research\\UMichigan\\Education_Teaching_Curricula\\2015_2016\\HS_853_Fall_2015\\Modules_docx\\data\\03_GraphNetwork_TermDocMatrix.rdata")
<center><b>Labeled graph</b> [[Image:SMHS_SciVisualization40.png|400px]] </center>
<center><b>Adjacency matrix</b> [[Image:SMHS_SciVisualization41.png|400px]] Coordinates are 1-6.</center>
# inspect part of the matrix
termDocMatrix [5:10,1:20]
# change it to a Boolean matrix == incidence matrix
termDocMatrix [termDocMatrix >=1] <- 1
# transform into a term-term adjacency matrix (n×n), where (i,j)th entries correspond to the number of edges
# node from xi to node xj.
# Matrix Multiplication in R: http://www.statmethods.net/advstats/matrix.html, dim(t(termDocMatrix))
termMatrix <- termDocMatrix %*% t(termDocMatrix)
# A graph has no loops, when all entries of the adjacency matrix on the main diagonal of are zeroes
# http://mathonline.wikidot.com/adjacency-matrices
diag(termMatrix)
# The matrix product of incidence matrix (B) and it’s transpose B×B^T represents the degrees of all nodes!
# inspect terms numbered 5 to 10
termMatrix[5:10,5:10]
# library("igraph")
# build a graph from the adjacency matrix
g <- graph.adjacency(termMatrix, weighted=T, mode="undirected")
plot(g)
# remove loops
g <- simplify(g); plot(g)
# set labels and degrees of V(g)
V(g)$\$$label <- V(g)$\$$name
V(g)$\$$degree <- degree(g)
# set seed to make the layout reproducible set.seed(1953) layout1 <- layout.fruchterman.reingold(g) # Fruchterman-Reingold layout plot(g, layout=layout1)
# plot(g, layout=layout.kamada.kawai) # tkplot(g, layout=layout.kamada.kawai)
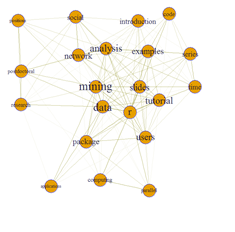
# Finesse the graph appearance – vertices and edges
V(g)$\$$label.cex <- 2.2 * V(g)$\$$degree / max(V(g)$\$$degree)+ .2
V(g)$\$$label.color <- rgb(0, 0, .2, .8)
V(g)$\$$frame.color <- rgb(0,0,1)
egam <- (log(E(g)$\$$weight)+.4) / max(log(E(g)$\$$weight)+.4)
E(g)$\$$color <- rgb(.5, .5, 0, egam) # Graph Edges, E(g)
E(g)$\$$width <- egam
# plot the graph in layout1
plot(g, layout=layout1)
V(g)$\$$label <- V(g)$\$$name
V(g)$\$$label.color <- rgb(0, 0, 0, 0.5)
V(g)$\$$label.dist <- 1.0 # relative distance of labels from node center
V(g)$\$$label.angle<- 3/8 #in radians
V(g)$\$$label.cex <- 1.4*V(g)$\$$degree/max(V(g)$\$$degree) + 1
V(g)$\$$color <- rgb(1, 0, 0, .4)
V(g)$\$$size <- 22 * V(g)$\$$degree / max(V(g)$\$$degree)+ 2
V(g)$\$$shape <- "rectangle"
# V(g)$\$$.size=10*(strwidth(V(g)$\$$label) + strwidth("oo")) * 10
# V(g)$\$$.size2=strheight("I") * 10
V(g)$\$$frame.color <- NA
# set vertex labels and their colors and sizes
# set edge width and color
E(g)$\$$width <- .3
E(g)$\$$color <- rgb(.5, .5, 0, .3)
set.seed(1234) plot(g, layout=layout.fruchterman.reingold)

Hands-on activity (oncological primary doctor and a second-opinion)
We use some of the Stanford Real Graph Data (http://snap.stanford.edu/data/) or this graph data on cancer patients seen by a primary doctor and a second-opinion doctor, stage of the disease, and diagnostic agreement between primary and secondary oncologist: Primary, Secondary, Stage, DxAgreement
# 03_GraphData_Health.txt
healthGraphTable <- read.table("https://umich.instructure.com/files/554234/download?download_frd=1", sep='\t', dec=',', header=T)
#specify the path, separator(tab, comma, ...), decimal point symbol, etc.
| Primary | Secondary | Stage | DxAgreement | |
| 1 | AA | DD | 3 | Y |
| 2 | AB | DD | 3 | R |
| 3 | AF | BA | 3 | Q |
| 4 | DD | DA | 3 | Q |
| 5 | CD | EC | 3 | X |
| 6 | DD | CE | 3 | Y |
# Transform the table into the required graph format: healthGraph.network<-graph.data.frame(healthGraphTable, directed=F) # the 'directed' attribute specifies whether the edges are directed # or equivalent irrespective of the position (1st vs 2nd column). For directed graphs use 'directed=T'
# Inspect the data:
V(healthGraph.network)
# prints the list of vertices (physicians/oncologists)
E(healthGraph.network)
# prints the list of edges (primary-secondary relationships)
degree(healthGraph.network)
# print the number of edges (relationships) per node (physician)
# first plot of graph plot(healthGraph.network)
#Subset the data. If we want to exclude only physicians who are mostly outside of the network # i.e., participate only tangentially (with 1 or 2 relationships only) # we can exclude nodes by subsetting the graph on the basis of the node/physician’s 'degree': healthGraph.out.network <- V(healthGraph.network)[degree(healthGraph.network)<=2] #identify those vertices part of less than or equal to 2 connections (edges) healthGraph.in.network <- delete.vertices(healthGraph.network, healthGraph.out.network) #exclude them from the graph
# Plot the data by specifying certain details about the graph, e.g., separate some nodes (people) by color:
V(healthGraph.in.network)$\$$color <- ifelse(V(healthGraph.in.network)$\$$name=='CA', 'blue', 'red')
#useful for highlighting certain people. Works by matching the name attribute of the vertex to the one specified in the 'ifelse' expression
# We can also color the connecting edges differently depending on the 'Stage':
E(healthGraph.in.network)$\$$color<-ifelse(E(healthGraph.in.network)$\$$Stage>3, "red", "grey")
# or depending on the different diagnostic agreement labels ('DxAgreement'):
E(healthGraph.in.network)$\$$color<-ifelse(E(healthGraph.in.network)$\$$DxAgreement =='X', "red", ifelse(E(healthGraph.in.network)$\$$DxAgreement=='Y', "blue", "grey"))
# Note: the example uses nested ifelse expressions which can be improved
# Additional attributes like size can be further specified in an analogous manner:
V(healthGraph.in.network)$\$$size<-degree(healthGraph.in.network)/10
#here the size of the vertices is specified by the degree of the vertex, so that people supervising more have get proportionally bigger dots. Getting the right scale gets some playing around with the parameters of the scale function (from the 'base' package)
# Note that if the same attribute is specified beforehand and inside the function, the former will be overridden.
# And finally the plot itself:
par(mai=c(0,0,1,0))
#this specifies the size of the margins, default settings leave too much free space on all sides
plot(healthGraph.in.network, #the graph to be plotted
layout=layout.fruchterman.reingold, # the layout method. see the igraph documentation for details
main='Onco Physician Network Example', #specifies the title
vertex.label.dist=0.5, #puts the name labels slightly off the dots
vertex.frame.color='blue', #the color of the border of the dots
vertex.label.color='black', #the color of the name labels
vertex.label.font=2, #the font of the name labels
vertex.label=V(healthGraph.in.network)$\$$name, #specifies the labels of the vertices
vertex.label.cex=1 #specifies the size of the font of the labels
)
# Save or export the plot as a metafile to the clipboard, a pdf or png (and other formats).
png(filename="org_network.png", height=1900, width=1200) #call the png writer
# alternatively print to high-res PDF file # pdf(file="org_network.pdf")
#run the plot
dev.off() #don’t forget to close the device
==='"`UNIQ--h-19--QINU`"'Pathway analysis===
Pathway analysis is a technique that reduces complexity and increased explanatory power in studies examining underlying biological structure of differentially expressed genes and proteins.
# install package
# install.packages("dendsort") – contains the “” dataset
# Data: Sample data matrix from the integrated pathway analysis of gastric cancer from the
# Cancer Genome Atlas (TCGA) study. A multivariate table obtained from the integrated pathway analysis
# of gastric cancer from the Cancer Genome Atlas (TCGA) study. Each column represents a pathway
# <u><b>consisting of a set of genes and each row represents a cohort of samples based on specific clinical
# or genetic features.</b></u> For each pair of a pathway and a feature, a continuous value of between
# 1 and -1 is assigned to score positive or negative association, respectively.
# A data frame with <u><b>215</b></u> rows and <u><b>117 variables</b></u>
library("dendsort")
data(sample_tcga)
dataTable <- t(sample_tcga)
head(dataTable)
write.csv(dataTable, "E:\\Ivo.dir\\Research\\UMichigan\\Education_Teaching_Curricula\\2015_2016\\HS_853_Fall_2015\\Modules_docx\\data\\03_TCGA_Data_117x215.csv")
# data.new <- read.csv("https://umich.instructure.com/files/330393/download?download_frd=1")
# install SPIA package: http://bioconductor.org/packages/2.6/bioc/html/SPIA.html
# source("http://bioconductor.org/biocLite.R")
# biocLite("SPIA")
library("SPIA")
# “top” Colorectal cancer dataset provided by SPIA package.
data(Vessels)
head(top)
# pathway analysis based on combined evidence;
# use nB=2000 or more for more accurate results
res<-spia(de=DE_Vessels,all=ALL_Vessels,organism="hsa",nB=500,plots=FALSE,beta=NULL,verbose=FALSE)
#make the output fit this screen
res$\$$Name=substr(res$\$$Name,1,10)
#show first 15 pathways, omitting KEGG links
res[1:15,-12]
GIS/Distortion mapping
# install the R GISTools package
# install.packages("GISTools")
library("GISTools")
data(georgia)
…
Java Applet: http://www.socr.ucla.edu/htmls/SOCR_Cartograhy.html Activities: http://wiki.stat.ucla.edu/socr/index.php/SOCR_Cartography_Project
Circos Connectogram/Table visualization
Circular chord/ribbon diagrams present a mechanism to visualize numeric tables containing information of directional relations. This type of chart visualizes tables in a circular way. Sectors of the plot is union(rownames(mat), colnames(mat)). When there is no rowname or colname, the chart assigns names for it (rows could be auto-named as "R1", "R2", ... and columns may be named as "C1", "C2").
• Circos: http://circos.ca
• R circlize: http://cran.r-project.org/web/packages/circlize/circlize.pdf