Difference between revisions of "SMHS BigDataBigSci GCM"
| Line 225: | Line 225: | ||
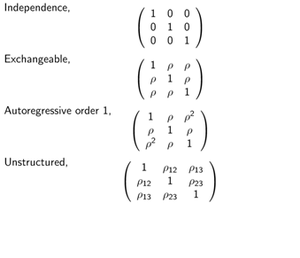
GEE is applied to handle correlated discrete and continuous outcome variables. For the outcome variables, it only requires specification of the first 2 moments and correlation among them. The goal is to estimate fixed parameters without specifying their joint distribution. The correlation is specified by one of these 4 alternatives (which is specified in the R call: geeglm(outcome ~ center + treat + sex + baseline + age, data = respiratory, family = "binomial", id = id, <b>corstr = " exchangeable"</b>, scale.fix = TRUE): | GEE is applied to handle correlated discrete and continuous outcome variables. For the outcome variables, it only requires specification of the first 2 moments and correlation among them. The goal is to estimate fixed parameters without specifying their joint distribution. The correlation is specified by one of these 4 alternatives (which is specified in the R call: geeglm(outcome ~ center + treat + sex + baseline + age, data = respiratory, family = "binomial", id = id, <b>corstr = " exchangeable"</b>, scale.fix = TRUE): | ||
| + | [[Image:SMHS_BigDataBigSci8.png|300px]] | ||
| + | ===Respiratory Illness GEE R example=== | ||
| + | This example is based on a data set on respiratory illness and the <b>geepack</b> package. The data is from a clinical study of the treatment effects on patients with respiratory illness. N=111 patients from 2 clinical centers randomized to receive either placebo or active treatments. 4 temporal examinations assessed the <b>respiratory state</b> of patients as good (=1) or poor (=0). Explanatory variables characterizing a patient were: <b>center</b> (1,2), treatment (A=active, P=placebo), <b>sex</b> (M=male, F=female), <b>age</b> (in years) at baseline. The values of the covariates were constant for the repeated elementary observations on each patient. | ||
| + | |||
| + | <b>Table 1</b> shows the number of patients for the response patterns across the 4 visits split by baseline-status and treatment. Baseline respiratory status = 0 appear to have either low or high number of positive responses. Baseline respiratory status = 1 tend to respond positively. <b>Table 2</b> describes the distribution of the number of positive responses per patient for sex and center. | ||
Revision as of 08:09, 6 May 2016
Contents
Model-based Analytics - Growth Curve Models
Latent growth curve models may be used to analyze longitudinal or temporal data where the outcome measure is assessed on multiple occasions, and we examine its change over time, e.g., the trajectory over time can be modeled as a linear or quadratic function. Random effects are used to capture individual differences by conveniently representing (continuous) latent variables, aka growth factors. To fit a linear growth model we may specify a model with two latent variables: a random intercept, and a random slope:
#load data 05_PPMI_top_UPDRS_Integrated_LongFormat.csv ( dim(myData) 661 71), wide
# setwd("/dir/")
myData <- read.csv("https://umich.instructure.com/files/330395/download?download_frd=1&verifier=v6jBvV4x94ka3EYcGKuXXg5BZNaOLBVp0xkJih0H",header=TRUE)
attach(myData)
# dichotomize the "ResearchGroup" variable table(myData$\$$ResearchGroup) myData$\$$ResearchGroup <- ifelse(myData$\$$ResearchGroup == "Control", 1, 0)
# linear growth model with 4 timepoints # intercept (i) and slope (s) with fixed coefficients # i =~ 1*t1 + 1*t2 + 1*t3 + 1*t4 (intercept/constant) # s =~ 0*t1 + 1*t2 + 2*t3 + 3*t4 (slope/linear term) # ??? =~ 0*t1 + 1*t2 + 2*t3 + 3*t4 (quadratic term)
In this model, we have fixed all the coefficients of the linear growth functions:
model4 <- ' i =~ 1*UPDRS_Part_I_Summary_Score_Baseline + 1*UPDRS_Part_I_Summary_Score_Month_03 + 1*UPDRS_Part_I_Summary_Score_Month_06 + 1*UPDRS_Part_I_Summary_Score_Month_09 + 1*UPDRS_Part_I_Summary_Score_Month_12 + 1*UPDRS_Part_I_Summary_Score_Month_18 + 1*UPDRS_Part_I_Summary_Score_Month_24 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Baseline + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_09 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_12 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24 + 1*UPDRS_Part_III_Summary_Score_Baseline + 1*UPDRS_Part_III_Summary_Score_Month_03 + 1*UPDRS_Part_III_Summary_Score_Month_06 + 1*UPDRS_Part_III_Summary_Score_Month_09 + 1*UPDRS_Part_III_Summary_Score_Month_12 + 1*UPDRS_Part_III_Summary_Score_Month_18 + 1*UPDRS_Part_III_Summary_Score_Month_24 + 1*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Baseline + 1*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06 + 1*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12 + 1*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_24 + 1*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline + 1*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06 + 1*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_12 + 1*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24 s =~ 0*UPDRS_Part_I_Summary_Score_Baseline + 1*UPDRS_Part_I_Summary_Score_Month_03 + 2*UPDRS_Part_I_Summary_Score_Month_06 + 3*UPDRS_Part_I_Summary_Score_Month_09 + 4*UPDRS_Part_I_Summary_Score_Month_12 + 5*UPDRS_Part_I_Summary_Score_Month_18 + 6*UPDRS_Part_I_Summary_Score_Month_24 + 0*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Baseline + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03 + 2*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06 + 3*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_09 + 4*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_12 + 5*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18 + 6*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24 + 0*UPDRS_Part_III_Summary_Score_Baseline + 1*UPDRS_Part_III_Summary_Score_Month_03 + 2*UPDRS_Part_III_Summary_Score_Month_06 + 3*UPDRS_Part_III_Summary_Score_Month_09 + 4*UPDRS_Part_III_Summary_Score_Month_12 + 5*UPDRS_Part_III_Summary_Score_Month_18 + 6*UPDRS_Part_III_Summary_Score_Month_24 + 0*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Baseline + 2*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06 + 4*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12 + 6*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_24 + 0*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline + 2*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06 + 4*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_12 + 6*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24 '
fit4 <- growth(model4, data=myData) summary(fit4) parameterEstimates(fit4) # extracts the values of the estimated parameters, the standard errors, # the z-values, the standardized parameter values, and returns a data frame fitted(fit4) # return the model-implied (fitted) covariance matrix (and mean vector) of a fitted model
# resid() function return (unstandardized) residuals of a fitted model including the difference between # the observed and implied covariance matrix and mean vector resid(fit4)
Measures of model quality (Comparative Fit Index (CFI), Root Mean Square Error of Approximation (RMSEA))
# report the fit measures as a signature vector: Comparative Fit Index (CFI), Root Mean Square Error of
# Approximation (RMSEA)
fitMeasures(fit4, c("cfi", "rmsea", "srmr"))
Comparative Fit Index (CFI) is an incremental measure directly based on the non-centrality measure. If d = χ2(df) where df are the degrees of freedom of the model, the Comparative Fit Index is:
FIX THIS!!!!!!!!!!!!!!!!(d(Null Model) - d(Proposed Model))/(d(Null Model)).
0≤CFI≤1 (by definition). It is interpreted as:
- CFI<0.9 - model fitting is poor.
- 0.9≤CFI≤0.95 is considered marginal,
- CFI>0.95 is good.
CFI is a relative index of model fit – it compare the fit of your model to the fit of (the worst) fitting null model.
Root Mean Square Error of Approximation (RMSEA) - “Ramsey”
An absolute measure of fit based on the non-centrality parameter: FIX EQUATION!!!!>√((χ2 - df)/(df×(N - 1))) ,
where N the sample size and df the degrees of freedom of the model. If χ2 < df, then the RMSEA∶=0. It has a penalty for complexity via the chi square to df ratio. The RMSEA is a popular measure of model fit.
- RMSEA < 0.01, excellent,
- RMSEA < 0.05, good
- RMSEA > 0.10 cutoff for poor fitting models
Standardized Root Mean Square Residual (SRMR) is an absolute measure of fit defined as the standardized difference between the observed correlation and the predicted correlation. A value of zero indicates perfect fit. The SRMR has no penalty for model complexity. SRMR <0.08 is considered a good fit.
# inspect the model results (report parameter table) inspect(fit4)
#install.packages("semTools")
# library("semTools")
A Simpler Model (fit5)
model5 <- '
# intercept and slope with fixed coefficients
i =~ UPDRS_Part_I_Summary_Score_Baseline + UPDRS_Part_I_Summary_Score_Month_03 + UPDRS_Part_I_Summary_Score_Month_24
s =~ 0*UPDRS_Part_I_Summary_Score_Baseline + 1*UPDRS_Part_I_Summary_Score_Month_03 + 6*UPDRS_Part_I_Summary_Score_Month_24
# regressions
i ~ R_fusiform_gyrus_Volume + Weight + ResearchGroup + Age + chr12_rs34637584_GT
s ~ R_fusiform_gyrus_Volume + Weight + ResearchGroup + Age + chr12_rs34637584_GT
# time-varying covariates
UPDRS_Part_I_Summary_Score_Baseline ~ Weight
UPDRS_Part_I_Summary_Score_Month_03 ~ ResearchGroup
UPDRS_Part_I_Summary_Score_Month_24 ~ Age
'
fit5 <- growth(model5, data=myData)
summary(fit5); fitMeasures(fit5, c("cfi", "rmsea", "srmr"))
parameterEstimates(fit5) # extracts the values of the estimated parameters, the standard errors,
# the z-values, the standardized parameter values, and returns a data frame
lavaan (0.5-18) converged normally after 99 iterations
Number of observations 661
Estimator ML
Minimum Function Test Statistic 3.703
Degrees of freedom 1
P-value (Chi-square) 0.054
Parameter estimates:
Information Expected
Standard Errors Standard
Estimate Std.err Z-value P(>|z|)
Latent variables:
i =~
UPDRS_P_I_S_S 1.000
UPDRS_P_I_S_S 1.074
UPDRS_P_I_S_S 1.172
s =~
UPDRS_P_I_S_S 0.000
UPDRS_P_I_S_S 1.000
UPDRS_P_I_S_S 6.000
Regressions: i ~ R_fsfrm_gyr_V 0.000 Weight 0.003 ResearchGroup -0.880 Age -0.009 c12_34637584_ -0.907 s ~ R_fsfrm_gyr_V -0.000 Weight -0.000 ResearchGroup -0.084 Age 0.002 c12_34637584_ -0.047 UPDRS_Part_I_Summary_Score_Baseline ~ Weight -0.000 UPDRS_Part_I_Summary_Score_Month_03 ~ ResearchGroup 0.693 UPDRS_Part_I_Summary_Score_Month_24 ~ Age -0.002
Covariances: i ~~ s 0.074
Intercepts: UPDRS_P_I_S_S 0.000 UPDRS_P_I_S_S 0.000 UPDRS_P_I_S_S 0.000 i 1.633 s -0.023
Variances: UPDRS_P_I_S_S 1.017 UPDRS_P_I_S_S 1.093 UPDRS_P_I_S_S 2.993 i 1.019 s -0.025
cfi rmsea srmr 0.996 0.064 0.008
fitted(fit5) # return the model-implied (fitted) covariance matrix (and mean vector) of a fitted model # write.table(fitted(fit5), file="C:\\Users\\Dinov\\Desktop\\test1.txt")
# resid() function return (unstandardized) residuals of a fitted model including the difference between # the observed and implied covariance matrix and mean vector resid(fit5)
# report the fit measures as a signature vector
fitMeasures(fit5, c("cfi", "rmsea", "srmr")) # comparative fit index (CFI)
# inspect the model results (report parameter table) inspect(fit5)
Note: See discussion of SEM modeling pros/cons.
Generalized Estimating Equation (GEE) Modeling
Generalized Estimating Equations (GEE) modeling is used for analyzing data with the following characteristics: (1) the observations within a group may be correlated, (2) observations in separate clusters are independent, (3) a monotone transformation of the expectation is linearly related to the explanatory variables, and (4) the variance is a function of the expectation. The expectation (#3) and the variance (# 4) are conditional given group-level or individual-level covariates.
GEE is applied to handle correlated discrete and continuous outcome variables. For the outcome variables, it only requires specification of the first 2 moments and correlation among them. The goal is to estimate fixed parameters without specifying their joint distribution. The correlation is specified by one of these 4 alternatives (which is specified in the R call: geeglm(outcome ~ center + treat + sex + baseline + age, data = respiratory, family = "binomial", id = id, corstr = " exchangeable", scale.fix = TRUE):
Respiratory Illness GEE R example
This example is based on a data set on respiratory illness and the geepack package. The data is from a clinical study of the treatment effects on patients with respiratory illness. N=111 patients from 2 clinical centers randomized to receive either placebo or active treatments. 4 temporal examinations assessed the respiratory state of patients as good (=1) or poor (=0). Explanatory variables characterizing a patient were: center (1,2), treatment (A=active, P=placebo), sex (M=male, F=female), age (in years) at baseline. The values of the covariates were constant for the repeated elementary observations on each patient.
Table 1 shows the number of patients for the response patterns across the 4 visits split by baseline-status and treatment. Baseline respiratory status = 0 appear to have either low or high number of positive responses. Baseline respiratory status = 1 tend to respond positively. Table 2 describes the distribution of the number of positive responses per patient for sex and center.
See also
- Back to Model-based Analytics
- Structural Equation Modeling (SEM)
- Next Section: Generalized Estimating Equation (GEE) Modeling
- SOCR Home page: http://www.socr.umich.edu
Translate this page: