Difference between revisions of "SOCR Videos Dashboard"
(→Reference/Citation) |
(→Protocol) |
||
| (16 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
The [http://socr.umich.edu/HTML5/Dashboard/ SOCR Dashboard] is a webapp providing a mechanism to integrate dispersed multi-source data and service the mashed information via human and machine interfaces in a secure, scalable manner. The Dashboard enables the exploration of subtle associations between variables, population strata, or clusters of data elements, which may be opaque to standard independent inspection of the individual sources. This a new platform includes a device agnostic tool for graphical querying, navigating and exploring the multivariate associations in complex heterogeneous datasets. | The [http://socr.umich.edu/HTML5/Dashboard/ SOCR Dashboard] is a webapp providing a mechanism to integrate dispersed multi-source data and service the mashed information via human and machine interfaces in a secure, scalable manner. The Dashboard enables the exploration of subtle associations between variables, population strata, or clusters of data elements, which may be opaque to standard independent inspection of the individual sources. This a new platform includes a device agnostic tool for graphical querying, navigating and exploring the multivariate associations in complex heterogeneous datasets. | ||
| − | ===Video=== | + | As an integrated Big-Data analytics suite, the SOCR Analytics Dashboard is designed for web-based merging, integration, and analysis of large, heterogeneous datasets. The package features a non-relational database for storage and integration of heterogeneous datasets, a JavaScript-based web server built using node.js, and a sleek front-end for visualization and real-time manipulation of data. |
| − | * | + | |
| + | ===Video Tutorial=== | ||
| + | * [https://www.youtube.com/embed/EDTDek1TqrU SOCR Data Dashboard Video Tutorial]. | ||
* [http://socr.umich.edu/docs/uploads/SOCR_Dashboard_Demo_Obesity.mkv SOCR Dashboard demonstration using user-specified data (in this case, multi-source pediatric obesity data)]. | * [http://socr.umich.edu/docs/uploads/SOCR_Dashboard_Demo_Obesity.mkv SOCR Dashboard demonstration using user-specified data (in this case, multi-source pediatric obesity data)]. | ||
| − | ===Script/Story Board=== | + | ====Script/Story Board==== |
(Time: concepts) | (Time: concepts) | ||
* Beginning: 2 different versions of webapp | * Beginning: 2 different versions of webapp | ||
| Line 20: | Line 22: | ||
=== Test Data=== | === Test Data=== | ||
| − | The following | + | The following datasets are used in this specific SOCR Data Dashboard demonstration: |
| − | * [http://socr.umich.edu/docs/uploads/MPOWER_Pilot.csv pediatric obesity dataset], | + | * [http://socr.umich.edu/docs/uploads/MPOWER_Pilot.csv pediatric obesity dataset], |
| − | * [http://socr.umich.edu/docs/uploads/PPMI_GSA_FAM_IntegratedDerived.csv Parkinson's disease neuroimaging-phenotypic-genetics dataset] | + | * [http://socr.umich.edu/docs/uploads/PPMI_GSA_FAM_IntegratedDerived.csv Parkinson's disease neuroimaging-phenotypic-genetics dataset], |
| + | * A [https://github.com/ini-bdds/bdbag BDBag/BagIt/ZIP formatted] version of the above 2 datasets is also [http://socr.umich.edu/docs/uploads/DataDashboard_TestData.zip available here]. | ||
<center>[[Image:SOCR_Videos_Dashboard_Fig1.png|500px]]</center> | <center>[[Image:SOCR_Videos_Dashboard_Fig1.png|500px]]</center> | ||
| + | |||
| + | ===Setting up the Dashboard=== | ||
| + | 1. Ensure the [https://nodejs.org/en/ node framework] has been installed. | ||
| + | |||
| + | 2. Install [https://docs.mongodb.com/manual/installation/ MongoDB]. This is not required, but if you want to see the demo using the default CMS, BLS, and Census data you need MongoDB installed. | ||
| + | |||
| + | 3. In a command prompt navigate to the ''server/mongodb/bin'' folder, and run mongod: | ||
| + | mongod --dbpath ../data | ||
| + | |||
| + | 4. Navigate to the root directory of the project in node, and run | ||
| + | npm install | ||
| + | |||
| + | 5. Once dependencies are installed, launch node server: | ||
| + | npm start | ||
| + | |||
| + | 6. Navigate to selected address in browser to launch webapp (default is [http://localhost:3000/ http://localhost:3000/]). | ||
| + | |||
| + | |||
| + | ===Example Case-Study I: Autism Brain Imaging Data Exchange (ABIDE)=== | ||
| + | This example describes a step-by-step recipe for using the Data Dashboard service to interrogate the [http://fcon_1000.projects.nitrc.org/indi/abide/ ABIDE data archive]. | ||
| + | |||
| + | The Autism Brain Imaging Data Exchange (ABIDE) data includes complex multi-source, incomplete, incongruent and multi-modal data for Autism spectrum disorder (ASD) pediatric cases. Core characteristics of ASD include impairment in social reciprocity, and by repetitive, restricted, and stereotyped behaviors/interests. | ||
| + | |||
| + | These data allow us to peak into the heterogeneous clinical, biological and physio-social characteristics of the ASD phenotypes. Specific biomedical questions that can be addressed via such data include early determination, tracking, diagnosis and prognosis, as well as assessing the clinical impact of alternative treatments. | ||
| + | |||
| + | The Data Dashboard webapp allows us to navigate this very heterogeneous data (n=1,112, k>2,000), graphically explore deep multivariate relations, associations, and trends. In addition, the dashboard service may be used to generate testable research hypothesis about various ASD traits that can be subsequently evaluated using prospective datasets. | ||
| + | |||
| + | ====Protocol==== | ||
| + | |||
| + | # Download the [http://socr.umich.edu/data/ABIDE_Data.zip BDDS ABIDE BagIt data object]. | ||
| + | # Extract the 4 data files (''neuroimaging_metrics.csv'', ''neuroimaging_metrics_metadata.csv'', ''neuroimaging_metrics_qc.csv'', ''phenotypes.csv''), which contain the relevant (appropriately named) components of the ABIDE dataset | ||
| + | # Start the [http://socr.umich.edu/HTML5/Dashboard/ Data Dashboard app] | ||
| + | # Select Free—Anchored Data | ||
| + | # Enter a new anchor variable subject_ID. [[Image:SOCR_Videos_Dashboard_Fig2.png|500px]] | ||
| + | # Import -- Phenotypes data (Pheno) | ||
| + | # Select a variable of interest (e.g., Sex) | ||
| + | # Add -- Chart (e.g., Pie Chart for Sex) from the Pheno data (Variable Sex) | ||
| + | # Add -- Chart (e.g., Histogram Chart for BMI) from the Pheno data (Variable BMI) – note that BMI has lots of missing data!!! | ||
| + | # Add -- Chart (e.g., Pie Chart of DX_Group or DSM_IV_TR) from the Pheno data | ||
| + | # Add -- Chart (e.g., Scatter Plot: FIQ vs. ADOS_COMM) | ||
| + | # Manipulate some of the charts (e.g., select females, BMIs, etc.) to examine the change in the other features (e.g., FIQ score). [[Image:SOCR_Videos_Dashboard_Fig3.png|500px]] | ||
| + | # Import -- Neuroimaging Data (NI), be patient, it will take some time to load the data. After the dialogue box shows up, click “Import” | ||
| + | # Add -- NI -- Select a variable of interest (e.g., Volume or SurfArea, as an example of a feature) click “Continue” | ||
| + | # Add -- NI -- Select a variable of interest (e.g., Fold_Ind, as an example of a feature) click “Continue” | ||
| + | # Add -- NI -- Select a variable of interest (e.g., Primary_Lobe, as an example of a feature) click “Continue” | ||
| + | # Manipulate some of the charts to study the impact on the other plots. [[Image:SOCR_Videos_Dashboard_Fig4.png|500px]]. | ||
| + | |||
| + | |||
| + | ===Example Case-Study 2: University of Michigan Udall Center for Excellence in Parkinson's Disease (Udall)=== | ||
| + | This example shows a step-by-step Data Dashboard protocol to interrogate a complex data archive containing [http://socr.umich.edu/docs/uploads/PPMI_GSA_FAM_IntegratedDerived.csv Parkinson's disease neuroimaging-phenotypic-genetics data]. This manuscript provides additional details about the dataset, specific clinical challenges, and examples of predictive analytics: [http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0157077 DOI: 10.1371/journal.pone.0157077]. | ||
| + | |||
| + | The data includes the following '''data elements''': | ||
| + | * FID_IID; L_insular_cortex_AvgMeanCurvature; L_insular_cortex_ComputeArea; L_insular_cortex_Volume; L_insular_cortex_ShapeIndex; L_insular_cortex_Curvedness; R_insular_cortex_AvgMeanCurvature; R_insular_cortex_ComputeArea; R_insular_cortex_Volume; R_insular_cortex_ShapeIndex; R_insular_cortex_Curvedness; L_cingulate_gyrus_AvgMeanCurvature; L_cingulate_gyrus_ComputeArea; L_cingulate_gyrus_Volume; L_cingulate_gyrus_ShapeIndex; L_cingulate_gyrus_Curvedness; R_cingulate_gyrus_AvgMeanCurvature; R_cingulate_gyrus_ComputeArea; R_cingulate_gyrus_Volume; R_cingulate_gyrus_ShapeIndex; R_cingulate_gyrus_Curvedness; L_caudate_AvgMeanCurvature; L_caudate_ComputeArea; L_caudate_Volume; L_caudate_ShapeIndex; L_caudate_Curvedness; R_caudate_AvgMeanCurvature; R_caudate_ComputeArea; R_caudate_Volume; R_caudate_ShapeIndex; R_caudate_Curvedness; L_putamen_AvgMeanCurvature; L_putamen_ComputeArea; L_putamen_Volume; L_putamen_ShapeIndex; L_putamen_Curvedness; R_putamen_AvgMeanCurvature; R_putamen_ComputeArea; R_putamen_Volume; R_putamen_ShapeIndex; R_putamen_Curvedness; L_hippocampus_AvgMeanCurvature; L_hippocampus_ComputeArea; L_hippocampus_Volume; L_hippocampus_ShapeIndex; L_hippocampus_Curvedness; R_hippocampus_AvgMeanCurvature; R_hippocampus_ComputeArea; R_hippocampus_Volume; R_hippocampus_ShapeIndex; R_hippocampus_Curvedness; cerebellum_AvgMeanCurvature; cerebellum_ComputeArea; cerebellum_Volume; cerebellum_ShapeIndex; cerebellum_Curvedness; brainstem_AvgMeanCurvature; brainstem_ComputeArea; brainstem_Volume; brainstem_ShapeIndex; brainstem_Curvedness; L_superior_frontal_gyrus_AvgMeanCurvature; L_superior_frontal_gyrus_ComputeArea; L_superior_frontal_gyrus_Volume; L_superior_frontal_gyrus_ShapeIndex; L_superior_frontal_gyrus_Curvedness; R_superior_frontal_gyrus_AvgMeanCurvature; R_superior_frontal_gyrus_ComputeArea; R_superior_frontal_gyrus_Volume; R_superior_frontal_gyrus_ShapeIndex; R_superior_frontal_gyrus_Curvedness; L_middle_frontal_gyrus_AvgMeanCurvature; L_middle_frontal_gyrus_ComputeArea; L_middle_frontal_gyrus_Volume; L_middle_frontal_gyrus_ShapeIndex; L_middle_frontal_gyrus_Curvedness; R_middle_frontal_gyrus_AvgMeanCurvature; R_middle_frontal_gyrus_ComputeArea; R_middle_frontal_gyrus_Volume; R_middle_frontal_gyrus_ShapeIndex; R_middle_frontal_gyrus_Curvedness; L_inferior_frontal_gyrus_AvgMeanCurvature; L_inferior_frontal_gyrus_ComputeArea; L_inferior_frontal_gyrus_Volume; L_inferior_frontal_gyrus_ShapeIndex; L_inferior_frontal_gyrus_Curvedness; R_inferior_frontal_gyrus_AvgMeanCurvature; R_inferior_frontal_gyrus_ComputeArea; R_inferior_frontal_gyrus_Volume; R_inferior_frontal_gyrus_ShapeIndex; R_inferior_frontal_gyrus_Curvedness; L_precentral_gyrus_AvgMeanCurvature; L_precentral_gyrus_ComputeArea; L_precentral_gyrus_Volume; L_precentral_gyrus_ShapeIndex; L_precentral_gyrus_Curvedness; R_precentral_gyrus_AvgMeanCurvature; R_precentral_gyrus_ComputeArea; R_precentral_gyrus_Volume; R_precentral_gyrus_ShapeIndex; R_precentral_gyrus_Curvedness; L_middle_orbitofrontal_gyrus_AvgMeanCurvature; L_middle_orbitofrontal_gyrus_ComputeArea; L_middle_orbitofrontal_gyrus_Volume; L_middle_orbitofrontal_gyrus_ShapeIndex; L_middle_orbitofrontal_gyrus_Curvedness; R_middle_orbitofrontal_gyrus_AvgMeanCurvature; R_middle_orbitofrontal_gyrus_ComputeArea; R_middle_orbitofrontal_gyrus_Volume; R_middle_orbitofrontal_gyrus_ShapeIndex; R_middle_orbitofrontal_gyrus_Curvedness; L_lateral_orbitofrontal_gyrus_AvgMeanCurvature; L_lateral_orbitofrontal_gyrus_ComputeArea; L_lateral_orbitofrontal_gyrus_Volume; L_lateral_orbitofrontal_gyrus_ShapeIndex; L_lateral_orbitofrontal_gyrus_Curvedness; R_lateral_orbitofrontal_gyrus_AvgMeanCurvature; R_lateral_orbitofrontal_gyrus_ComputeArea; R_lateral_orbitofrontal_gyrus_Volume; R_lateral_orbitofrontal_gyrus_ShapeIndex; R_lateral_orbitofrontal_gyrus_Curvedness; L_gyrus_rectus_AvgMeanCurvature; L_gyrus_rectus_ComputeArea; L_gyrus_rectus_Volume; L_gyrus_rectus_ShapeIndex; L_gyrus_rectus_Curvedness; R_gyrus_rectus_AvgMeanCurvature; R_gyrus_rectus_ComputeArea; R_gyrus_rectus_Volume; R_gyrus_rectus_ShapeIndex; R_gyrus_rectus_Curvedness; L_postcentral_gyrus_AvgMeanCurvature; L_postcentral_gyrus_ComputeArea; L_postcentral_gyrus_Volume; L_postcentral_gyrus_ShapeIndex; L_postcentral_gyrus_Curvedness; R_postcentral_gyrus_AvgMeanCurvature; R_postcentral_gyrus_ComputeArea; R_postcentral_gyrus_Volume; R_postcentral_gyrus_ShapeIndex; R_postcentral_gyrus_Curvedness; L_superior_parietal_gyrus_AvgMeanCurvature; L_superior_parietal_gyrus_ComputeArea; L_superior_parietal_gyrus_Volume; L_superior_parietal_gyrus_ShapeIndex; L_superior_parietal_gyrus_Curvedness; R_superior_parietal_gyrus_AvgMeanCurvature; R_superior_parietal_gyrus_ComputeArea; R_superior_parietal_gyrus_Volume; R_superior_parietal_gyrus_ShapeIndex; R_superior_parietal_gyrus_Curvedness; L_supramarginal_gyrus_AvgMeanCurvature; L_supramarginal_gyrus_ComputeArea; L_supramarginal_gyrus_Volume; L_supramarginal_gyrus_ShapeIndex; L_supramarginal_gyrus_Curvedness; R_supramarginal_gyrus_AvgMeanCurvature; R_supramarginal_gyrus_ComputeArea; R_supramarginal_gyrus_Volume; R_supramarginal_gyrus_ShapeIndex; R_supramarginal_gyrus_Curvedness; L_angular_gyrus_AvgMeanCurvature; L_angular_gyrus_ComputeArea; L_angular_gyrus_Volume; L_angular_gyrus_ShapeIndex; L_angular_gyrus_Curvedness; R_angular_gyrus_AvgMeanCurvature; R_angular_gyrus_ComputeArea; R_angular_gyrus_Volume; R_angular_gyrus_ShapeIndex; R_angular_gyrus_Curvedness; L_precuneus_AvgMeanCurvature; L_precuneus_ComputeArea; L_precuneus_Volume; L_precuneus_ShapeIndex; L_precuneus_Curvedness; R_precuneus_AvgMeanCurvature; R_precuneus_ComputeArea; R_precuneus_Volume; R_precuneus_ShapeIndex; R_precuneus_Curvedness; L_superior_occipital_gyrus_AvgMeanCurvature; L_superior_occipital_gyrus_ComputeArea; L_superior_occipital_gyrus_Volume; L_superior_occipital_gyrus_ShapeIndex; L_superior_occipital_gyrus_Curvedness; R_superior_occipital_gyrus_AvgMeanCurvature; R_superior_occipital_gyrus_ComputeArea; R_superior_occipital_gyrus_Volume; R_superior_occipital_gyrus_ShapeIndex; R_superior_occipital_gyrus_Curvedness; L_middle_occipital_gyrus_AvgMeanCurvature; L_middle_occipital_gyrus_ComputeArea; L_middle_occipital_gyrus_Volume; L_middle_occipital_gyrus_ShapeIndex; L_middle_occipital_gyrus_Curvedness; R_middle_occipital_gyrus_AvgMeanCurvature; R_middle_occipital_gyrus_ComputeArea; R_middle_occipital_gyrus_Volume; R_middle_occipital_gyrus_ShapeIndex; R_middle_occipital_gyrus_Curvedness; L_inferior_occipital_gyrus_AvgMeanCurvature; L_inferior_occipital_gyrus_ComputeArea; L_inferior_occipital_gyrus_Volume; L_inferior_occipital_gyrus_ShapeIndex; L_inferior_occipital_gyrus_Curvedness; R_inferior_occipital_gyrus_AvgMeanCurvature; R_inferior_occipital_gyrus_ComputeArea; R_inferior_occipital_gyrus_Volume; R_inferior_occipital_gyrus_ShapeIndex; R_inferior_occipital_gyrus_Curvedness; L_cuneus_AvgMeanCurvature; L_cuneus_ComputeArea; L_cuneus_Volume; L_cuneus_ShapeIndex; L_cuneus_Curvedness; R_cuneus_AvgMeanCurvature; R_cuneus_ComputeArea; R_cuneus_Volume; R_cuneus_ShapeIndex; R_cuneus_Curvedness; L_superior_temporal_gyrus_AvgMeanCurvature; L_superior_temporal_gyrus_ComputeArea; L_superior_temporal_gyrus_Volume; L_superior_temporal_gyrus_ShapeIndex; L_superior_temporal_gyrus_Curvedness; R_superior_temporal_gyrus_AvgMeanCurvature; R_superior_temporal_gyrus_ComputeArea; R_superior_temporal_gyrus_Volume; R_superior_temporal_gyrus_ShapeIndex; R_superior_temporal_gyrus_Curvedness; L_middle_temporal_gyrus_AvgMeanCurvature; L_middle_temporal_gyrus_ComputeArea; L_middle_temporal_gyrus_Volume; L_middle_temporal_gyrus_ShapeIndex; L_middle_temporal_gyrus_Curvedness; R_middle_temporal_gyrus_AvgMeanCurvature; R_middle_temporal_gyrus_ComputeArea; R_middle_temporal_gyrus_Volume; R_middle_temporal_gyrus_ShapeIndex; R_middle_temporal_gyrus_Curvedness; L_inferior_temporal_gyrus_AvgMeanCurvature; L_inferior_temporal_gyrus_ComputeArea; L_inferior_temporal_gyrus_Volume; L_inferior_temporal_gyrus_ShapeIndex; L_inferior_temporal_gyrus_Curvedness; R_inferior_temporal_gyrus_AvgMeanCurvature; R_inferior_temporal_gyrus_ComputeArea; R_inferior_temporal_gyrus_Volume; R_inferior_temporal_gyrus_ShapeIndex; R_inferior_temporal_gyrus_Curvedness; L_parahippocampal_gyrus_AvgMeanCurvature; L_parahippocampal_gyrus_ComputeArea; L_parahippocampal_gyrus_Volume; L_parahippocampal_gyrus_ShapeIndex; L_parahippocampal_gyrus_Curvedness; R_parahippocampal_gyrus_AvgMeanCurvature; R_parahippocampal_gyrus_ComputeArea; R_parahippocampal_gyrus_Volume; R_parahippocampal_gyrus_ShapeIndex; R_parahippocampal_gyrus_Curvedness; L_lingual_gyrus_AvgMeanCurvature; L_lingual_gyrus_ComputeArea; L_lingual_gyrus_Volume; L_lingual_gyrus_ShapeIndex; L_lingual_gyrus_Curvedness; R_lingual_gyrus_AvgMeanCurvature; R_lingual_gyrus_ComputeArea; R_lingual_gyrus_Volume; R_lingual_gyrus_ShapeIndex; R_lingual_gyrus_Curvedness; L_fusiform_gyrus_AvgMeanCurvature; L_fusiform_gyrus_ComputeArea; L_fusiform_gyrus_Volume; L_fusiform_gyrus_ShapeIndex; L_fusiform_gyrus_Curvedness; R_fusiform_gyrus_AvgMeanCurvature; R_fusiform_gyrus_ComputeArea; R_fusiform_gyrus_Volume; R_fusiform_gyrus_ShapeIndex; R_fusiform_gyrus_Curvedness; FID(father); FID(mother); Sex; Phenotype; Sex; Weight; ResearchGroup; VisitID; Age. | ||
| + | |||
| + | These data represent multi-source, incomplete, incongruent and multi-modal information a large cohort of of volunteers (PD, SWEDD, and Control). The data allow us to peak into the disease heterogeneity of PD phenotypes, ask specific health related questions about diagnosis, tracking, and prognosis, as well as about assessing the clinical impact of alternative treatments. | ||
| + | |||
| + | The Data Dashboard webapp allows us to explore these very heterogeneous data (n=576, k>289, longitudinal), graphically explore deep multivariate relations, associations, and trends, as well as generate testable research hypothesis about various PD traits that can be subsequently evaluated using prospective datasets. | ||
| + | |||
| + | ====Protocol==== | ||
| + | |||
| + | # Download locally the [http://socr.umich.edu/docs/uploads/PPMI_GSA_FAM_IntegratedDerived.csv PD data (CSV)]. | ||
| + | # Start the [http://socr.umich.edu/HTML5/Dashboard/ Data Dashboard App] | ||
| + | # Select '''Free—Anchored''' Data | ||
| + | # Enter the following in the pop-up dialog box: | ||
| + | ## Cohort variable name: '''Age''' | ||
| + | ## Minimum cohort variable value: '''30''' | ||
| + | ## Maximum cohort variable value: '''85''' | ||
| + | ## Number of Bins: '''10''' | ||
| + | # Click '''Start''' (to initialize the Dashboard) | ||
| + | # Click '''Import''' (to load the [http://socr.umich.edu/docs/uploads/PPMI_GSA_FAM_IntegratedDerived.csv PD data (CSV)] from your computer) | ||
| + | # Click '''Browse''' (to select the local CSV file), under '''Enter Source Name''', enter '''PD''', and then Click '''Continue''' | ||
| + | # When the '''Custom Dataset''' dialog pop-up shows, Click '''Import''' (wait a few seconds for the data to be ingested into the Dashboard) | ||
| + | # Now you are ready to '''Add''' charts to the Dashboard Canvas. | ||
| + | # Click '''Add''' (+) chart and when the '''Add New Widget''' dialog (add chart) shows up, select ''PPMI_GSA_FAM_IntegratedDerived.csv'' (data source) and ''Age'' (Select the data variable), and Click '''Continue''' | ||
| + | # On the '''Select Chart Type''' dialog, choose ''histogram'' from the list of charts and then Click '''Continue'''. You should see the first plot re[resenting a histogram distribution of Ages in the PD cohort | ||
| + | # Next we will repeat the same process several more times to add more charts and plots to the collage of graphs in the Dashboard. | ||
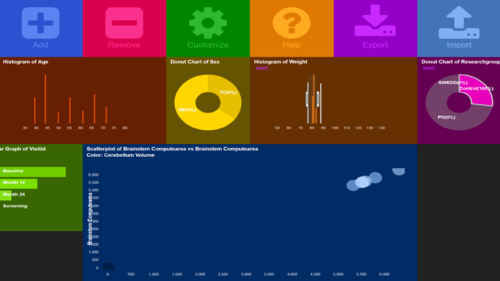
| + | # For instance, we can add '''Sex''' (Pie chart), '''Weight''' (histogram), '''ResearchGroup''' (Pie Chart), '''VisitID''' (Bar chart), and '''Brainstem computearea''' vs. '''Cerebellum volume''' (Scatterplot) | ||
| + | # At this point, the ''Dashboard canvas'' should look something like the image shown below | ||
| + | <center>[[Image:SOCR_Videos_Dashboard_Fig24.png|500px]]</center> | ||
| + | # We are now ready to initiate the process of (1) visual database query and exploratory analytics, and (2) dynamic visualization of intricate multivariate patterns that can be used to formulate new (testable) research hypotheses | ||
| + | # Over each chart, using the mouse, you can (1) select a sub-cohort by Clicking on specific chart components, or (2) Choose a range of values (phenotypes) by Right-clicking-and-dragging | ||
| + | # In this dynamic exploration, mind the changes the occur on all other charts in response to the manual graphical stratification of the database | ||
| + | # On each chart, Clicking the '''Reset''' text in the top-left corner removes any prior user stratification based on the specific features included in the chart | ||
| + | # The result of one specific data stratification is illustrated on the figure below. | ||
| + | <center>[[Image:SOCR_Videos_Dashboard_Fig25.png|500px]]</center> | ||
| + | |||
| + | A very important feature of the SOCR Data Dashboard is that complies with data privacy protection and regulatory guidelines (e.g., HIPAA) to protect personal information. The Dashboard models the distribution in the entire data archive and simulates from the joint distribution to avoid rendering individualized data. As such, the Data Dashboard is neither an exact representation of the data nor it guarantees that a match/no-match found indicates the presence/absence of a specific participant from the study population. | ||
| + | |||
| + | ===Reference Materials=== | ||
| + | * BagIt/BDBag Data File Format: | ||
| + | ** [https://github.com/ini-bdds/bdbag Source code] and [https://github.com/ini-bdds/minid Format] | ||
| + | ** [http://bd2k.ini.usc.edu/assets/all-hands-meeting/minid_v0.1_Nov_2015.pdf Docs (PDF)] | ||
| + | * Data Dashboard | ||
| + | ** [https://github.com/SOCR/SOCR-Dashboard Source code] | ||
| + | ** [[SOCR_Videos_Dashboard#Video_Tutorial | Videos]] | ||
| + | ** [http://socr.umich.edu/HTML5/Dashboard/ Service/App] | ||
=== Reference/Citation=== | === Reference/Citation=== | ||
Latest revision as of 13:57, 17 August 2018
Contents
- 1 SOCR Videos: SOCR Data Dashboard Videos
SOCR Videos: SOCR Data Dashboard Videos
The SOCR Dashboard is a webapp providing a mechanism to integrate dispersed multi-source data and service the mashed information via human and machine interfaces in a secure, scalable manner. The Dashboard enables the exploration of subtle associations between variables, population strata, or clusters of data elements, which may be opaque to standard independent inspection of the individual sources. This a new platform includes a device agnostic tool for graphical querying, navigating and exploring the multivariate associations in complex heterogeneous datasets.
As an integrated Big-Data analytics suite, the SOCR Analytics Dashboard is designed for web-based merging, integration, and analysis of large, heterogeneous datasets. The package features a non-relational database for storage and integration of heterogeneous datasets, a JavaScript-based web server built using node.js, and a sleek front-end for visualization and real-time manipulation of data.
Video Tutorial
- SOCR Data Dashboard Video Tutorial.
- SOCR Dashboard demonstration using user-specified data (in this case, multi-source pediatric obesity data).
Script/Story Board
(Time: concepts)
- Beginning: 2 different versions of webapp
- 1:22: Sample file contains many omissions and errors
- 1:46: Variable list auto-populated
- 2:20: Completely different dataset
- 2:45: Much larger data size
- 3:30: Cross-filtering functionality
- 3:47: Dynamic tile-based layout
- 4:30: Context sensitive menus
- 7:00: Custom Datasets
Test Data
The following datasets are used in this specific SOCR Data Dashboard demonstration:
- pediatric obesity dataset,
- Parkinson's disease neuroimaging-phenotypic-genetics dataset,
- A BDBag/BagIt/ZIP formatted version of the above 2 datasets is also available here.

Setting up the Dashboard
1. Ensure the node framework has been installed.
2. Install MongoDB. This is not required, but if you want to see the demo using the default CMS, BLS, and Census data you need MongoDB installed.
3. In a command prompt navigate to the server/mongodb/bin folder, and run mongod:
mongod --dbpath ../data
4. Navigate to the root directory of the project in node, and run
npm install
5. Once dependencies are installed, launch node server:
npm start
6. Navigate to selected address in browser to launch webapp (default is http://localhost:3000/).
Example Case-Study I: Autism Brain Imaging Data Exchange (ABIDE)
This example describes a step-by-step recipe for using the Data Dashboard service to interrogate the ABIDE data archive.
The Autism Brain Imaging Data Exchange (ABIDE) data includes complex multi-source, incomplete, incongruent and multi-modal data for Autism spectrum disorder (ASD) pediatric cases. Core characteristics of ASD include impairment in social reciprocity, and by repetitive, restricted, and stereotyped behaviors/interests.
These data allow us to peak into the heterogeneous clinical, biological and physio-social characteristics of the ASD phenotypes. Specific biomedical questions that can be addressed via such data include early determination, tracking, diagnosis and prognosis, as well as assessing the clinical impact of alternative treatments.
The Data Dashboard webapp allows us to navigate this very heterogeneous data (n=1,112, k>2,000), graphically explore deep multivariate relations, associations, and trends. In addition, the dashboard service may be used to generate testable research hypothesis about various ASD traits that can be subsequently evaluated using prospective datasets.
Protocol
- Download the BDDS ABIDE BagIt data object.
- Extract the 4 data files (neuroimaging_metrics.csv, neuroimaging_metrics_metadata.csv, neuroimaging_metrics_qc.csv, phenotypes.csv), which contain the relevant (appropriately named) components of the ABIDE dataset
- Start the Data Dashboard app
- Select Free—Anchored Data
- Enter a new anchor variable subject_ID.
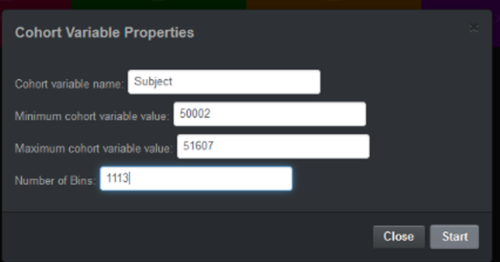
- Import -- Phenotypes data (Pheno)
- Select a variable of interest (e.g., Sex)
- Add -- Chart (e.g., Pie Chart for Sex) from the Pheno data (Variable Sex)
- Add -- Chart (e.g., Histogram Chart for BMI) from the Pheno data (Variable BMI) – note that BMI has lots of missing data!!!
- Add -- Chart (e.g., Pie Chart of DX_Group or DSM_IV_TR) from the Pheno data
- Add -- Chart (e.g., Scatter Plot: FIQ vs. ADOS_COMM)
- Manipulate some of the charts (e.g., select females, BMIs, etc.) to examine the change in the other features (e.g., FIQ score).
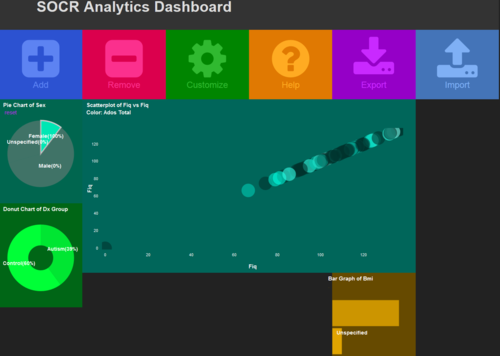
- Import -- Neuroimaging Data (NI), be patient, it will take some time to load the data. After the dialogue box shows up, click “Import”
- Add -- NI -- Select a variable of interest (e.g., Volume or SurfArea, as an example of a feature) click “Continue”
- Add -- NI -- Select a variable of interest (e.g., Fold_Ind, as an example of a feature) click “Continue”
- Add -- NI -- Select a variable of interest (e.g., Primary_Lobe, as an example of a feature) click “Continue”
- Manipulate some of the charts to study the impact on the other plots.
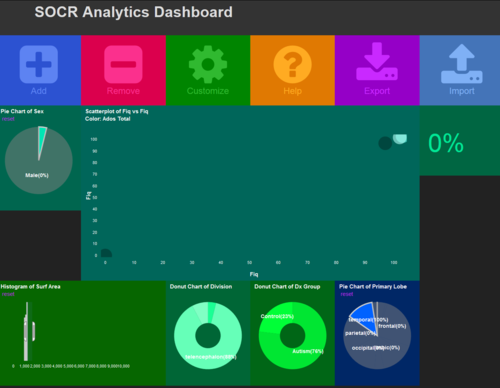 .
.
Example Case-Study 2: University of Michigan Udall Center for Excellence in Parkinson's Disease (Udall)
This example shows a step-by-step Data Dashboard protocol to interrogate a complex data archive containing Parkinson's disease neuroimaging-phenotypic-genetics data. This manuscript provides additional details about the dataset, specific clinical challenges, and examples of predictive analytics: DOI: 10.1371/journal.pone.0157077.
The data includes the following data elements:
- FID_IID; L_insular_cortex_AvgMeanCurvature; L_insular_cortex_ComputeArea; L_insular_cortex_Volume; L_insular_cortex_ShapeIndex; L_insular_cortex_Curvedness; R_insular_cortex_AvgMeanCurvature; R_insular_cortex_ComputeArea; R_insular_cortex_Volume; R_insular_cortex_ShapeIndex; R_insular_cortex_Curvedness; L_cingulate_gyrus_AvgMeanCurvature; L_cingulate_gyrus_ComputeArea; L_cingulate_gyrus_Volume; L_cingulate_gyrus_ShapeIndex; L_cingulate_gyrus_Curvedness; R_cingulate_gyrus_AvgMeanCurvature; R_cingulate_gyrus_ComputeArea; R_cingulate_gyrus_Volume; R_cingulate_gyrus_ShapeIndex; R_cingulate_gyrus_Curvedness; L_caudate_AvgMeanCurvature; L_caudate_ComputeArea; L_caudate_Volume; L_caudate_ShapeIndex; L_caudate_Curvedness; R_caudate_AvgMeanCurvature; R_caudate_ComputeArea; R_caudate_Volume; R_caudate_ShapeIndex; R_caudate_Curvedness; L_putamen_AvgMeanCurvature; L_putamen_ComputeArea; L_putamen_Volume; L_putamen_ShapeIndex; L_putamen_Curvedness; R_putamen_AvgMeanCurvature; R_putamen_ComputeArea; R_putamen_Volume; R_putamen_ShapeIndex; R_putamen_Curvedness; L_hippocampus_AvgMeanCurvature; L_hippocampus_ComputeArea; L_hippocampus_Volume; L_hippocampus_ShapeIndex; L_hippocampus_Curvedness; R_hippocampus_AvgMeanCurvature; R_hippocampus_ComputeArea; R_hippocampus_Volume; R_hippocampus_ShapeIndex; R_hippocampus_Curvedness; cerebellum_AvgMeanCurvature; cerebellum_ComputeArea; cerebellum_Volume; cerebellum_ShapeIndex; cerebellum_Curvedness; brainstem_AvgMeanCurvature; brainstem_ComputeArea; brainstem_Volume; brainstem_ShapeIndex; brainstem_Curvedness; L_superior_frontal_gyrus_AvgMeanCurvature; L_superior_frontal_gyrus_ComputeArea; L_superior_frontal_gyrus_Volume; L_superior_frontal_gyrus_ShapeIndex; L_superior_frontal_gyrus_Curvedness; R_superior_frontal_gyrus_AvgMeanCurvature; R_superior_frontal_gyrus_ComputeArea; R_superior_frontal_gyrus_Volume; R_superior_frontal_gyrus_ShapeIndex; R_superior_frontal_gyrus_Curvedness; L_middle_frontal_gyrus_AvgMeanCurvature; L_middle_frontal_gyrus_ComputeArea; L_middle_frontal_gyrus_Volume; L_middle_frontal_gyrus_ShapeIndex; L_middle_frontal_gyrus_Curvedness; R_middle_frontal_gyrus_AvgMeanCurvature; R_middle_frontal_gyrus_ComputeArea; R_middle_frontal_gyrus_Volume; R_middle_frontal_gyrus_ShapeIndex; R_middle_frontal_gyrus_Curvedness; L_inferior_frontal_gyrus_AvgMeanCurvature; L_inferior_frontal_gyrus_ComputeArea; L_inferior_frontal_gyrus_Volume; L_inferior_frontal_gyrus_ShapeIndex; L_inferior_frontal_gyrus_Curvedness; R_inferior_frontal_gyrus_AvgMeanCurvature; R_inferior_frontal_gyrus_ComputeArea; R_inferior_frontal_gyrus_Volume; R_inferior_frontal_gyrus_ShapeIndex; R_inferior_frontal_gyrus_Curvedness; L_precentral_gyrus_AvgMeanCurvature; L_precentral_gyrus_ComputeArea; L_precentral_gyrus_Volume; L_precentral_gyrus_ShapeIndex; L_precentral_gyrus_Curvedness; R_precentral_gyrus_AvgMeanCurvature; R_precentral_gyrus_ComputeArea; R_precentral_gyrus_Volume; R_precentral_gyrus_ShapeIndex; R_precentral_gyrus_Curvedness; L_middle_orbitofrontal_gyrus_AvgMeanCurvature; L_middle_orbitofrontal_gyrus_ComputeArea; L_middle_orbitofrontal_gyrus_Volume; L_middle_orbitofrontal_gyrus_ShapeIndex; L_middle_orbitofrontal_gyrus_Curvedness; R_middle_orbitofrontal_gyrus_AvgMeanCurvature; R_middle_orbitofrontal_gyrus_ComputeArea; R_middle_orbitofrontal_gyrus_Volume; R_middle_orbitofrontal_gyrus_ShapeIndex; R_middle_orbitofrontal_gyrus_Curvedness; L_lateral_orbitofrontal_gyrus_AvgMeanCurvature; L_lateral_orbitofrontal_gyrus_ComputeArea; L_lateral_orbitofrontal_gyrus_Volume; L_lateral_orbitofrontal_gyrus_ShapeIndex; L_lateral_orbitofrontal_gyrus_Curvedness; R_lateral_orbitofrontal_gyrus_AvgMeanCurvature; R_lateral_orbitofrontal_gyrus_ComputeArea; R_lateral_orbitofrontal_gyrus_Volume; R_lateral_orbitofrontal_gyrus_ShapeIndex; R_lateral_orbitofrontal_gyrus_Curvedness; L_gyrus_rectus_AvgMeanCurvature; L_gyrus_rectus_ComputeArea; L_gyrus_rectus_Volume; L_gyrus_rectus_ShapeIndex; L_gyrus_rectus_Curvedness; R_gyrus_rectus_AvgMeanCurvature; R_gyrus_rectus_ComputeArea; R_gyrus_rectus_Volume; R_gyrus_rectus_ShapeIndex; R_gyrus_rectus_Curvedness; L_postcentral_gyrus_AvgMeanCurvature; L_postcentral_gyrus_ComputeArea; L_postcentral_gyrus_Volume; L_postcentral_gyrus_ShapeIndex; L_postcentral_gyrus_Curvedness; R_postcentral_gyrus_AvgMeanCurvature; R_postcentral_gyrus_ComputeArea; R_postcentral_gyrus_Volume; R_postcentral_gyrus_ShapeIndex; R_postcentral_gyrus_Curvedness; L_superior_parietal_gyrus_AvgMeanCurvature; L_superior_parietal_gyrus_ComputeArea; L_superior_parietal_gyrus_Volume; L_superior_parietal_gyrus_ShapeIndex; L_superior_parietal_gyrus_Curvedness; R_superior_parietal_gyrus_AvgMeanCurvature; R_superior_parietal_gyrus_ComputeArea; R_superior_parietal_gyrus_Volume; R_superior_parietal_gyrus_ShapeIndex; R_superior_parietal_gyrus_Curvedness; L_supramarginal_gyrus_AvgMeanCurvature; L_supramarginal_gyrus_ComputeArea; L_supramarginal_gyrus_Volume; L_supramarginal_gyrus_ShapeIndex; L_supramarginal_gyrus_Curvedness; R_supramarginal_gyrus_AvgMeanCurvature; R_supramarginal_gyrus_ComputeArea; R_supramarginal_gyrus_Volume; R_supramarginal_gyrus_ShapeIndex; R_supramarginal_gyrus_Curvedness; L_angular_gyrus_AvgMeanCurvature; L_angular_gyrus_ComputeArea; L_angular_gyrus_Volume; L_angular_gyrus_ShapeIndex; L_angular_gyrus_Curvedness; R_angular_gyrus_AvgMeanCurvature; R_angular_gyrus_ComputeArea; R_angular_gyrus_Volume; R_angular_gyrus_ShapeIndex; R_angular_gyrus_Curvedness; L_precuneus_AvgMeanCurvature; L_precuneus_ComputeArea; L_precuneus_Volume; L_precuneus_ShapeIndex; L_precuneus_Curvedness; R_precuneus_AvgMeanCurvature; R_precuneus_ComputeArea; R_precuneus_Volume; R_precuneus_ShapeIndex; R_precuneus_Curvedness; L_superior_occipital_gyrus_AvgMeanCurvature; L_superior_occipital_gyrus_ComputeArea; L_superior_occipital_gyrus_Volume; L_superior_occipital_gyrus_ShapeIndex; L_superior_occipital_gyrus_Curvedness; R_superior_occipital_gyrus_AvgMeanCurvature; R_superior_occipital_gyrus_ComputeArea; R_superior_occipital_gyrus_Volume; R_superior_occipital_gyrus_ShapeIndex; R_superior_occipital_gyrus_Curvedness; L_middle_occipital_gyrus_AvgMeanCurvature; L_middle_occipital_gyrus_ComputeArea; L_middle_occipital_gyrus_Volume; L_middle_occipital_gyrus_ShapeIndex; L_middle_occipital_gyrus_Curvedness; R_middle_occipital_gyrus_AvgMeanCurvature; R_middle_occipital_gyrus_ComputeArea; R_middle_occipital_gyrus_Volume; R_middle_occipital_gyrus_ShapeIndex; R_middle_occipital_gyrus_Curvedness; L_inferior_occipital_gyrus_AvgMeanCurvature; L_inferior_occipital_gyrus_ComputeArea; L_inferior_occipital_gyrus_Volume; L_inferior_occipital_gyrus_ShapeIndex; L_inferior_occipital_gyrus_Curvedness; R_inferior_occipital_gyrus_AvgMeanCurvature; R_inferior_occipital_gyrus_ComputeArea; R_inferior_occipital_gyrus_Volume; R_inferior_occipital_gyrus_ShapeIndex; R_inferior_occipital_gyrus_Curvedness; L_cuneus_AvgMeanCurvature; L_cuneus_ComputeArea; L_cuneus_Volume; L_cuneus_ShapeIndex; L_cuneus_Curvedness; R_cuneus_AvgMeanCurvature; R_cuneus_ComputeArea; R_cuneus_Volume; R_cuneus_ShapeIndex; R_cuneus_Curvedness; L_superior_temporal_gyrus_AvgMeanCurvature; L_superior_temporal_gyrus_ComputeArea; L_superior_temporal_gyrus_Volume; L_superior_temporal_gyrus_ShapeIndex; L_superior_temporal_gyrus_Curvedness; R_superior_temporal_gyrus_AvgMeanCurvature; R_superior_temporal_gyrus_ComputeArea; R_superior_temporal_gyrus_Volume; R_superior_temporal_gyrus_ShapeIndex; R_superior_temporal_gyrus_Curvedness; L_middle_temporal_gyrus_AvgMeanCurvature; L_middle_temporal_gyrus_ComputeArea; L_middle_temporal_gyrus_Volume; L_middle_temporal_gyrus_ShapeIndex; L_middle_temporal_gyrus_Curvedness; R_middle_temporal_gyrus_AvgMeanCurvature; R_middle_temporal_gyrus_ComputeArea; R_middle_temporal_gyrus_Volume; R_middle_temporal_gyrus_ShapeIndex; R_middle_temporal_gyrus_Curvedness; L_inferior_temporal_gyrus_AvgMeanCurvature; L_inferior_temporal_gyrus_ComputeArea; L_inferior_temporal_gyrus_Volume; L_inferior_temporal_gyrus_ShapeIndex; L_inferior_temporal_gyrus_Curvedness; R_inferior_temporal_gyrus_AvgMeanCurvature; R_inferior_temporal_gyrus_ComputeArea; R_inferior_temporal_gyrus_Volume; R_inferior_temporal_gyrus_ShapeIndex; R_inferior_temporal_gyrus_Curvedness; L_parahippocampal_gyrus_AvgMeanCurvature; L_parahippocampal_gyrus_ComputeArea; L_parahippocampal_gyrus_Volume; L_parahippocampal_gyrus_ShapeIndex; L_parahippocampal_gyrus_Curvedness; R_parahippocampal_gyrus_AvgMeanCurvature; R_parahippocampal_gyrus_ComputeArea; R_parahippocampal_gyrus_Volume; R_parahippocampal_gyrus_ShapeIndex; R_parahippocampal_gyrus_Curvedness; L_lingual_gyrus_AvgMeanCurvature; L_lingual_gyrus_ComputeArea; L_lingual_gyrus_Volume; L_lingual_gyrus_ShapeIndex; L_lingual_gyrus_Curvedness; R_lingual_gyrus_AvgMeanCurvature; R_lingual_gyrus_ComputeArea; R_lingual_gyrus_Volume; R_lingual_gyrus_ShapeIndex; R_lingual_gyrus_Curvedness; L_fusiform_gyrus_AvgMeanCurvature; L_fusiform_gyrus_ComputeArea; L_fusiform_gyrus_Volume; L_fusiform_gyrus_ShapeIndex; L_fusiform_gyrus_Curvedness; R_fusiform_gyrus_AvgMeanCurvature; R_fusiform_gyrus_ComputeArea; R_fusiform_gyrus_Volume; R_fusiform_gyrus_ShapeIndex; R_fusiform_gyrus_Curvedness; FID(father); FID(mother); Sex; Phenotype; Sex; Weight; ResearchGroup; VisitID; Age.
These data represent multi-source, incomplete, incongruent and multi-modal information a large cohort of of volunteers (PD, SWEDD, and Control). The data allow us to peak into the disease heterogeneity of PD phenotypes, ask specific health related questions about diagnosis, tracking, and prognosis, as well as about assessing the clinical impact of alternative treatments.
The Data Dashboard webapp allows us to explore these very heterogeneous data (n=576, k>289, longitudinal), graphically explore deep multivariate relations, associations, and trends, as well as generate testable research hypothesis about various PD traits that can be subsequently evaluated using prospective datasets.
Protocol
- Download locally the PD data (CSV).
- Start the Data Dashboard App
- Select Free—Anchored Data
- Enter the following in the pop-up dialog box:
- Cohort variable name: Age
- Minimum cohort variable value: 30
- Maximum cohort variable value: 85
- Number of Bins: 10
- Click Start (to initialize the Dashboard)
- Click Import (to load the PD data (CSV) from your computer)
- Click Browse (to select the local CSV file), under Enter Source Name, enter PD, and then Click Continue
- When the Custom Dataset dialog pop-up shows, Click Import (wait a few seconds for the data to be ingested into the Dashboard)
- Now you are ready to Add charts to the Dashboard Canvas.
- Click Add (+) chart and when the Add New Widget dialog (add chart) shows up, select PPMI_GSA_FAM_IntegratedDerived.csv (data source) and Age (Select the data variable), and Click Continue
- On the Select Chart Type dialog, choose histogram from the list of charts and then Click Continue. You should see the first plot re[resenting a histogram distribution of Ages in the PD cohort
- Next we will repeat the same process several more times to add more charts and plots to the collage of graphs in the Dashboard.
- For instance, we can add Sex (Pie chart), Weight (histogram), ResearchGroup (Pie Chart), VisitID (Bar chart), and Brainstem computearea vs. Cerebellum volume (Scatterplot)
- At this point, the Dashboard canvas should look something like the image shown below
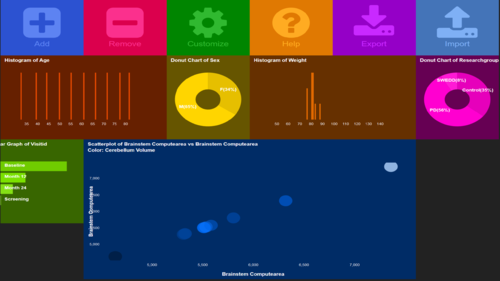
- We are now ready to initiate the process of (1) visual database query and exploratory analytics, and (2) dynamic visualization of intricate multivariate patterns that can be used to formulate new (testable) research hypotheses
- Over each chart, using the mouse, you can (1) select a sub-cohort by Clicking on specific chart components, or (2) Choose a range of values (phenotypes) by Right-clicking-and-dragging
- In this dynamic exploration, mind the changes the occur on all other charts in response to the manual graphical stratification of the database
- On each chart, Clicking the Reset text in the top-left corner removes any prior user stratification based on the specific features included in the chart
- The result of one specific data stratification is illustrated on the figure below.
A very important feature of the SOCR Data Dashboard is that complies with data privacy protection and regulatory guidelines (e.g., HIPAA) to protect personal information. The Dashboard models the distribution in the entire data archive and simulates from the joint distribution to avoid rendering individualized data. As such, the Data Dashboard is neither an exact representation of the data nor it guarantees that a match/no-match found indicates the presence/absence of a specific participant from the study population.
Reference Materials
- BagIt/BDBag Data File Format:
- Data Dashboard
Reference/Citation
- Husain, SS, Kalinin, A, Truong, A, Dinov, ID. (2015) SOCR data dashboard: an integrated big data archive mashing medicare, labor, census and econometric information. Journal of Big Data, 2(13):1-18. DOI: 10.1186/s40537-015-0018-z.
- Dashboard GitHub source code
- SOCR Dashboard Webapp
- SOCR Home page: http://www.socr.ucla.edu or http://www.StatisticsResource.org
Translate this page: