Difference between revisions of "NISER 081107 ID"
m |
m (Text replacement - "{{translate|pageName=http://wiki.stat.ucla.edu/socr/" to ""{{translate|pageName=http://wiki.socr.umich.edu/") |
||
| (4 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | == [http://wiki.stat.ucla.edu/niser NISER Wiki Resourses have Migrated to here!]== | + | == [http://wiki.stat.ucla.edu/niser/index.php/NISER_EduMaterials NISER Wiki Resourses have Migrated to here!]== |
| − | == [ | + | |
| + | == [http://wiki.stat.ucla.edu/niser/index.php/NISER_EduMaterials_GeneralActivities NISER General Activities] - Mercury Contamination in Fish: A multi-disciplinary Activity == | ||
==Summary== | ==Summary== | ||
| − | Visualization, understanding and interpreting real data may be challenging because of noise in the data, data complexity, multiple variables, hidden relations between variables and large variation. This [ | + | Visualization, understanding and interpreting real data may be challenging because of noise in the data, data complexity, multiple variables, hidden relations between variables and large variation. This [http://wiki.stat.ucla.edu/niser NISER] activity demonstrates how to use free Internet-based IT-tools and resources to solve problems that arise in the areas of biological, chemical, medical and social research. |
==Goals== | ==Goals== | ||
| − | This [ | + | This [http://wiki.stat.ucla.edu/niser NISER] activity has the following specific goals: |
* to demonstrate the typical research investigation pipeline - from problem formulation, to data collection, visualization, analysis and interpretation; | * to demonstrate the typical research investigation pipeline - from problem formulation, to data collection, visualization, analysis and interpretation; | ||
* to illustrate the variety of portable freely available Internet-based Java tools, computational resources and learning materials for solving practical problems; | * to illustrate the variety of portable freely available Internet-based Java tools, computational resources and learning materials for solving practical problems; | ||
| Line 30: | Line 31: | ||
==Methods & Approaches== | ==Methods & Approaches== | ||
| − | [[Image:NISER_081107_ID_Fig11.png|150px|thumbnail|right| [http://www.falstad.com/qmatom/ Atomic structure] | + | [[Image:NISER_081107_ID_Fig11.png|150px|thumbnail|right| [http://www.falstad.com/qmatom/ Atomic structure]]] |
We now discuss varieties of scientific methods, models, tools and strategies for data modeling, understanding, inference and visualization of the data in this specific driving biological problem, as well as, discuss checking and affirming underlying model or technique assumptions. | We now discuss varieties of scientific methods, models, tools and strategies for data modeling, understanding, inference and visualization of the data in this specific driving biological problem, as well as, discuss checking and affirming underlying model or technique assumptions. | ||
===Physics=== | ===Physics=== | ||
| − | * What is an atom and what is the atomic structure of mercury? | + | * What is an atom and what is the [http://environmentalchemistry.com/yogi/periodic/Hg.html#Atomic atomic structure of mercury]? |
| − | * | + | * [http://environmentalchemistry.com/yogi/periodic/Hg.html#Physical Physical Properties of mercury]. |
| − | + | * [http://environmentalchemistry.com/yogi/periodic/Hg-pg2.html#Nuclides Mercury nuclides and isotopes]. | |
| − | + | * How can [http://cobweb.ecn.purdue.edu/~epados/mercbuild/src/cleanup.htm small high-concentration mercury spills may be cleaned]? | |
| − | |||
| − | |||
| − | |||
| − | * | ||
===Biology=== | ===Biology=== | ||
| − | [[Image:NISER_081107_ID_Fig9.png|150px|thumbnail|right| [http://www.rcsb.org/pdb/explore.do?structureId=2O01 Chlorophyll 3D Structure] | + | [[Image:NISER_081107_ID_Fig9.png|150px|thumbnail|right| [http://www.rcsb.org/pdb/explore.do?structureId=2O01 Chlorophyll 3D Structure]]] |
* Why is mercury (and other [http://en.wikipedia.org/wiki/Heavy_metals heavy metals]) accumulating in muscle tissue? | * Why is mercury (and other [http://en.wikipedia.org/wiki/Heavy_metals heavy metals]) accumulating in muscle tissue? | ||
* What causes the [http://chem.sis.nlm.nih.gov/chemidplus/ProxyServlet?objectHandle=DBMaint&actionHandle=default&nextPage=jsp/chemidheavy/ResultScreen.jsp&ROW_NUM=0&TXTSUPERLISTID=007439976 toxicity of larger than normal amounts of mercury in the body]? | * What causes the [http://chem.sis.nlm.nih.gov/chemidplus/ProxyServlet?objectHandle=DBMaint&actionHandle=default&nextPage=jsp/chemidheavy/ResultScreen.jsp&ROW_NUM=0&TXTSUPERLISTID=007439976 toxicity of larger than normal amounts of mercury in the body]? | ||
* The [http://en.wikipedia.org/wiki/Chlorophyll Chlorophyll] molecule. | * The [http://en.wikipedia.org/wiki/Chlorophyll Chlorophyll] molecule. | ||
* [http://www.rcsb.org/pdb/explore.do?structureId=2O01 3D structure of Chlorophyll] using [http://www.rcsb.org/pdb PDB/JMol viewer]. | * [http://www.rcsb.org/pdb/explore.do?structureId=2O01 3D structure of Chlorophyll] using [http://www.rcsb.org/pdb PDB/JMol viewer]. | ||
| − | + | * [http://environmentalchemistry.com/yogi/periodic/Hg.html#Regulatory The biology and regulatory limits for mercury exposure]. | |
| − | + | * [http://en.wikipedia.org/wiki/Alkalinity Alkalinity] represents the ability of a solution to neutralize [http://en.wikipedia.org/wiki/Acid acids] to the equivalence point of carbonate or bicarbonate. Alkalinity is closely related to a solution's acid neutralizing capacity. However, the acid neutralizing capacity refers to the combination of the solution and solids present and the contribution of solids can dominate the acid neutralizing capacity. | |
| − | + | * Mercury uptake by plants | |
| − | * | + | ** Mercury uptake by plants is low (Jenkins, 1981; Paasivirta, 1991), however plants may be used for monitoring mercury contaminations. |
| + | ** Trees uptake mercury through the xylem, from soil via the root system or from direct deposition via foliage and bark. | ||
| + | * The Biology of mercury uptake: | ||
| + | ** In fish, mercury appears primarily as methylmercury and that's the most common form of mercury transfered through the food chain. | ||
| + | ** The most conducive environmental conditions for the methylation and uptake of mercury into fish include: '''low pH (<7), low alkalinity (<20 mg/L), low calcium (<15 mg/L), high total organic carbon (TOC), low chlorophyll-a (<l0 μμg/L)''', and significant seasonal fluctuations in water level (Rada et al., 1989; Lange et al., 1993; Wiener, 1995). | ||
| + | ** Methylmercury is produced initially by microbial activity, as it is insoluble in water, and forms colloids with humus, which in turn transfer mercury from soils to water. | ||
| + | ** Dissolved organic carbon complexes transport mercury. | ||
| + | ** As pH decreases, partitioning of mercury is shifted from sediment to water. | ||
| + | ** The major pathways for mercury removal from solution is chemical binding with reduced '''sulphur, which has a high affinity for mercury'''. | ||
| + | ** Increased concentrations of some metals, e.g., iron and manganese, affect the non-biological methylation of mercury. Acid volatile sulfides like iron and manganese sulfides are a reactive pool of solid phase sulfides are available to bind with metals, such as mercury (Di Toro et al., 1990), which makes mercury is less available for uptake by fish. | ||
===Ethotoxicology=== | ===Ethotoxicology=== | ||
| Line 58: | Line 64: | ||
===Chemistry=== | ===Chemistry=== | ||
| − | [[Image:NISER_081107_ID_Fig12.png|150px|thumbnail|right| [http://www.chemcollective.org/applets/pertable.php Periodic Table] | + | [[Image:NISER_081107_ID_Fig12.png|150px|thumbnail|right| [http://www.chemcollective.org/applets/pertable.php Periodic Table]]] |
* [http://www.dartmouth.edu/~chemlab/info/resources/p_table/Periodic.html Periodic Table] | * [http://www.dartmouth.edu/~chemlab/info/resources/p_table/Periodic.html Periodic Table] | ||
* [http://en.wikipedia.org/wiki/Alkalinity Alkalinity] | * [http://en.wikipedia.org/wiki/Alkalinity Alkalinity] | ||
* [http://en.wikipedia.org/wiki/PH pH] | * [http://en.wikipedia.org/wiki/PH pH] | ||
* See the position of '''Mercury''' (''Hg'') in the [http://www.chemcollective.org/applets/pertable.php interactive Periodic table] and discuss its chemical properties. | * See the position of '''Mercury''' (''Hg'') in the [http://www.chemcollective.org/applets/pertable.php interactive Periodic table] and discuss its chemical properties. | ||
| − | + | * [http://environmentalchemistry.com/yogi/periodic/Hg.html#Chemical Chemical properties of mercury]. | |
| − | + | * [http://environmentalchemistry.com/yogi/periodic/Hg-compounds.html#Compounds Common mercury chemical componds]. | |
| − | |||
| − | |||
| − | * | ||
===Engineering=== | ===Engineering=== | ||
| − | [[Image:NISER_081107_ID_Fig13.png|150px|thumbnail|right| Engineering Thumbnail TBD | + | [[Image:NISER_081107_ID_Fig13.png|150px|thumbnail|right| Engineering Thumbnail TBD]] |
| − | * | + | * [http://www.sg.ohio-state.edu/research/biotechnology/?ID=R/PS-1 Novel engineering approaches for mercury decontamination using specialized bacteria]. |
| − | + | * [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6VB5-4CHRHXY-3&_user=4423&_coverDate=09%2F30%2F2004&_rdoc=17&_fmt=summary&_orig=browse&_srch=doc-info(%23toc%235917%232004%23998689997%23509220%23FLA%23display%23Volume)&_cdi=5917&_sort=d&_docanchor=&view=c&_ct=17&_acct=C000059605&_version=1&_urlVersion=0&_userid=4423&md5=c0a7645b14f421c39f09307eee6f4744 Remediation and passive decontamination for mercury pollution by Capping, Dredging & Natural attenuation] | |
| − | + | * Mercury biomagnifies as it moves up trophic levels in the food chain. Biomagnification by plankton from water may be as high as 100,000 times. Food web enrichment is apparent in plankton and aquatic insects, insect-eating fish, piscivorous fish, and fish-eating birds (Paasivirta, 1991). | |
| − | |||
| − | |||
| − | * | ||
===Mathematics=== | ===Mathematics=== | ||
| − | [[Image:NISER_081107_ID_Fig14.png|150px|thumbnail|right| [http://socr.ucla.edu Math Modeling] | + | [[Image:NISER_081107_ID_Fig14.png|150px|thumbnail|right| [http://socr.ucla.edu Math Modeling]]] |
* Fit '''models''' to some of the variables in [[NISER_081107_ID_Data | this dataset]]. | * Fit '''models''' to some of the variables in [[NISER_081107_ID_Data | this dataset]]. | ||
| − | ** We can easily compute the histogram of the average (per lake) mercury measurements and fit in [http://socr.ucla.edu/htmls/SOCR_Distributions.html Normal], [http://socr.ucla.edu/htmls/SOCR_Distributions.html Exponential] or [ | + | ** We can easily compute the histogram of the average (per lake) mercury measurements and fit in [http://socr.ucla.edu/htmls/SOCR_Distributions.html Normal], [http://socr.ucla.edu/htmls/SOCR_Distributions.html Exponential] or [http://wiki.stat.ucla.edu/socr/index.php/About_pages_for_SOCR_Distributions other distribution models] to the histogram of the average mercury measurement using [http://socr.ucla.edu/htmls/SOCR_Charts.html SOCR Charts] and [http://www.socr.ucla.edu/htmls/SOCR_Modeler.html SOCR Modeler] (see [http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials examples here]). The left image below shows the fit of a <math>Normal (\mu=10.54, \sigma^2=45.64)</math> distribution model and the right image shows the (offset by 0.8) <math>Exponential(\lambda = 10.54)</math> model fit to the mercury histogram. The parameters in both cases are automatically calculated from the data using [http://en.wikipedia.org/wiki/Maximum_likelihood_estimation maximum likelihood estimation]. <center>[[Image:NISER_081107_ID_Fig3.png|300px]] [[Image:NISER_081107_ID_Fig4.png|300px]]</center> |
| − | ** We can also demonstrate the effect of using a wavelet decomposition of the column data (''avg_mercury''). The image below demonstrates the '''wavelet representation''' of this column vector data using Daubechies' 2 wavelet basis, where only the largest 10% of the wavelet coefficients were used to reconstruct/synthesize the (compressed) wavelet representation of the data. More information about [http://en.wikipedia.org/wiki/Wavelets wavelets] and [ | + | ** We can also demonstrate the effect of using a wavelet decomposition of the column data (''avg_mercury''). The image below demonstrates the '''wavelet representation''' of this column vector data using Daubechies' 2 wavelet basis, where only the largest 10% of the wavelet coefficients were used to reconstruct/synthesize the (compressed) wavelet representation of the data. More information about [http://en.wikipedia.org/wiki/Wavelets wavelets] and [http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials_GamesActivitiesWavelets wavelets-based activities may be found here]. You can use the [http://www.socr.ucla.edu/htmls/SOCR_Modeler.html SOCR Modeler] to fit is any of a number of wavelet models to data and observe the effect of wavelet shrinkage on the corresponding wavelet model fit. |
<center>[[Image:NISER_081107_ID_Fig7.png|400px]]</center> | <center>[[Image:NISER_081107_ID_Fig7.png|400px]]</center> | ||
===Statistics=== | ===Statistics=== | ||
| − | [[Image:NISER_081107_ID_Fig15.png|150px|thumbnail|right| [http://socr.ucla.edu Statistical Analysis] | + | [[Image:NISER_081107_ID_Fig15.png|150px|thumbnail|right| [http://socr.ucla.edu Statistical Analysis]]] |
* Are there associations in [[NISER_081107_ID_Data | these data]] between ''Alkalinity'', ''pH'', ''Calcium'' and ''Chlorophyll''? Using [http://www.socr.ucla.edu/htmls/SOCR_Analyses.html SOCR Analyses] construct regression plots illustrating the relations between ''Calcium, Chlorophyll'' and '' Avg_Mercury'', as shown on the image below. | * Are there associations in [[NISER_081107_ID_Data | these data]] between ''Alkalinity'', ''pH'', ''Calcium'' and ''Chlorophyll''? Using [http://www.socr.ucla.edu/htmls/SOCR_Analyses.html SOCR Analyses] construct regression plots illustrating the relations between ''Calcium, Chlorophyll'' and '' Avg_Mercury'', as shown on the image below. | ||
<center>[[Image:NISER_081107_ID_Fig2.png|400px]]</center> | <center>[[Image:NISER_081107_ID_Fig2.png|400px]]</center> | ||
* Generate some exploratory data analyses (EDA plots). For instance, the figure below demonstrates how you can use [http://www.socr.ucla.edu/htmls/SOCR_Charts.html SOCR Charts] to obtain the scatter plot of ''Alkalinity'' vs. ''pH''. What are some inferences about the bivariate relations between these variables? | * Generate some exploratory data analyses (EDA plots). For instance, the figure below demonstrates how you can use [http://www.socr.ucla.edu/htmls/SOCR_Charts.html SOCR Charts] to obtain the scatter plot of ''Alkalinity'' vs. ''pH''. What are some inferences about the bivariate relations between these variables? | ||
<center>[[Image:NISER_081107_ID_Fig1.png|400px]]</center> | <center>[[Image:NISER_081107_ID_Fig1.png|400px]]</center> | ||
| − | * What is the distribution of the average amount of ''mercury'' in the entire sample? We can use [http://www.socr.ucla.edu/htmls/SOCR_Charts.html SOCR Charts] to construct the histogram of the ''avg_mercury'' column data. To learn how to use [http://www.socr.ucla.edu/htmls/SOCR_Charts.html SOCR Charts] see these [ | + | * What is the distribution of the average amount of ''mercury'' in the entire sample? We can use [http://www.socr.ucla.edu/htmls/SOCR_Charts.html SOCR Charts] to construct the histogram of the ''avg_mercury'' column data. To learn how to use [http://www.socr.ucla.edu/htmls/SOCR_Charts.html SOCR Charts] see these [http://wiki.stat.ucla.edu/socr/index.php/SOCR_EduMaterials_ChartsActivities Charts Activities]. Notice the effect the bin-size parameter has on the shape of the data histogram (you can smoothly vary this parameter). Although, there are [http://en.wikipedia.org/wiki/Histogram many choices for determining the bin-size parameter], in general, the optimal choice of the histogram bin-size parameter may be determined by <math>{2\times(IQR)} \over {N^{1\over 3}}</math>, where the interquartile range <math>IQR= Q_3 - Q_1</math>, and <math>Q_3</math> and <math>Q_1</math> are the '''third''' and '''first''' [http://en.wikipedia.org/wiki/Quartiles quartiles] for the sample data, respectively. |
<center>[[Image:NISER_081107_ID_Fig8.png|400px]]</center> | <center>[[Image:NISER_081107_ID_Fig8.png|400px]]</center> | ||
* Is there evidence of statistical differences between the two groups (according to the ''age_data'' variable) in either ''Alkalinity'' or ''pH''? | * Is there evidence of statistical differences between the two groups (according to the ''age_data'' variable) in either ''Alkalinity'' or ''pH''? | ||
| Line 110: | Line 110: | ||
<hr> | <hr> | ||
| − | * | + | * NISER Home page: http://www.NSER.org |
| − | {{translate|pageName=http://wiki. | + | "{{translate|pageName=http://wiki.socr.umich.edu/index.php?title=NISER_081107_ID}} |
Latest revision as of 12:33, 3 March 2020
Contents
NISER Wiki Resourses have Migrated to here!
NISER General Activities - Mercury Contamination in Fish: A multi-disciplinary Activity
Summary
Visualization, understanding and interpreting real data may be challenging because of noise in the data, data complexity, multiple variables, hidden relations between variables and large variation. This NISER activity demonstrates how to use free Internet-based IT-tools and resources to solve problems that arise in the areas of biological, chemical, medical and social research.
Goals
This NISER activity has the following specific goals:
- to demonstrate the typical research investigation pipeline - from problem formulation, to data collection, visualization, analysis and interpretation;
- to illustrate the variety of portable freely available Internet-based Java tools, computational resources and learning materials for solving practical problems;
- to provide a hands-on example of interdisciplinary training, cross-over of research techniques, data, models and expertise to enhance contemporary science education;
- to promote interactions between different science education areas and stimulate the development of new and synergistic learning materials and course curricula across disciplines.
Motivation/Problem
Mercury contamination of edible freshwater fish poses a direct threat to human health. Largemouth bass is a fresh water fish that was studied in 53 different Florida lakes to examine the factors that influence the level of mercury contamination. Water samples were collected from the surface of the middle of each lake in August 1990 and then again in March 1991. The pH level, the amount of chlorophyll, calcium, and alkalinity were measured in each sample. Also, samples of fish were taken from each lake with sample sizes ranging from 4 to 44 fish. The age of each fish and mercury concentration in the muscle tissue was measured. Since fish absorb mercury over time, older fish will tend to have higher concentrations. Here is a detailed discussion of why mercury accumilates in muscle tissues and why is it toxic in larger amounts.
Data
Largemouth bass were studied in 53 different Florida lakes to examine the factors that influence the level of mercury contamination. Water samples were collected from the surface of the middle of each lake in August 1990 and then again in March 1991. The pH level, the amount of chlorophyll, calcium, and alkalinity were measured in each sample. The average of the August and March values were used in the analysis. Next, a sample of fish was taken from each lake with sample sizes ranging from 4 to 44 fish. The age of each fish and mercury concentration in the muscle tissue was measured.
Challenge
To make a fair comparison of the fish in different lakes using a regression estimate of the expected mercury concentration in a three-year-old fish as the standardized value for each lake. Determine the age of the individual fish in some lakes and correlate this with the average mercury concentration of the sampled fish.
Florida has set a standard of 1/2 part per million as the unsafe level of mercury concentration in edible foods. 45.3% of the lakes exceed this level. The smallest level of mercury concentration that the measuring instrument can detect is 40 parts per billion. Any level below that was set to 40 parts per billion. This, of course, "flattens out" the slope of the relationship at the low end as well as affecting the standardized values. These observations are usually on young fish.
Logarithmic transformations on some of the variables may provide insights into the relationships among the other variables in the study. For instance, alkalinity level may be associated with mercury concentration, and may help account for the higher levels of mercury.
Methods & Approaches
We now discuss varieties of scientific methods, models, tools and strategies for data modeling, understanding, inference and visualization of the data in this specific driving biological problem, as well as, discuss checking and affirming underlying model or technique assumptions.
Physics
- What is an atom and what is the atomic structure of mercury?
- Physical Properties of mercury.
- Mercury nuclides and isotopes.
- How can small high-concentration mercury spills may be cleaned?
Biology
- Why is mercury (and other heavy metals) accumulating in muscle tissue?
- What causes the toxicity of larger than normal amounts of mercury in the body?
- The Chlorophyll molecule.
- 3D structure of Chlorophyll using PDB/JMol viewer.
- The biology and regulatory limits for mercury exposure.
- Alkalinity represents the ability of a solution to neutralize acids to the equivalence point of carbonate or bicarbonate. Alkalinity is closely related to a solution's acid neutralizing capacity. However, the acid neutralizing capacity refers to the combination of the solution and solids present and the contribution of solids can dominate the acid neutralizing capacity.
- Mercury uptake by plants
- Mercury uptake by plants is low (Jenkins, 1981; Paasivirta, 1991), however plants may be used for monitoring mercury contaminations.
- Trees uptake mercury through the xylem, from soil via the root system or from direct deposition via foliage and bark.
- The Biology of mercury uptake:
- In fish, mercury appears primarily as methylmercury and that's the most common form of mercury transfered through the food chain.
- The most conducive environmental conditions for the methylation and uptake of mercury into fish include: low pH (<7), low alkalinity (<20 mg/L), low calcium (<15 mg/L), high total organic carbon (TOC), low chlorophyll-a (<l0 μμg/L), and significant seasonal fluctuations in water level (Rada et al., 1989; Lange et al., 1993; Wiener, 1995).
- Methylmercury is produced initially by microbial activity, as it is insoluble in water, and forms colloids with humus, which in turn transfer mercury from soils to water.
- Dissolved organic carbon complexes transport mercury.
- As pH decreases, partitioning of mercury is shifted from sediment to water.
- The major pathways for mercury removal from solution is chemical binding with reduced sulphur, which has a high affinity for mercury.
- Increased concentrations of some metals, e.g., iron and manganese, affect the non-biological methylation of mercury. Acid volatile sulfides like iron and manganese sulfides are a reactive pool of solid phase sulfides are available to bind with metals, such as mercury (Di Toro et al., 1990), which makes mercury is less available for uptake by fish.
Ethotoxicology
Ethotoxicology is the study of the behavioral consequences of exposure to toxic chemicals. Toxic chemicals, like heavy metals and estrogen mimicking compounds, may influence behavior. For instance, there are documented changes in how some animals respond to novel situations, vocalize, and expose themselves to the risk of predation as a function of chemical exposure. Thus, by quantifying time allocation using an event recorder (like JWatcher), and using various statistical analyses to analyze these data, it is often possible to have a behavioral indicator of chemical exposure. And, by quantifying such behaviors in a variety of species, and then using phylogenetic software (like Mesquite), it is possible to study the evolution of such behavioral effects. We are in the process of developing new applets and learning materials demonstrating the effects of toxic exposure to such compounds on animal behavior.
Chemistry
- Periodic Table
- Alkalinity
- pH
- See the position of Mercury (Hg) in the interactive Periodic table and discuss its chemical properties.
- Chemical properties of mercury.
- Common mercury chemical componds.
Engineering
- Novel engineering approaches for mercury decontamination using specialized bacteria.
- Remediation and passive decontamination for mercury pollution by Capping, Dredging & Natural attenuation
- Mercury biomagnifies as it moves up trophic levels in the food chain. Biomagnification by plankton from water may be as high as 100,000 times. Food web enrichment is apparent in plankton and aquatic insects, insect-eating fish, piscivorous fish, and fish-eating birds (Paasivirta, 1991).
Mathematics
- Fit models to some of the variables in this dataset.
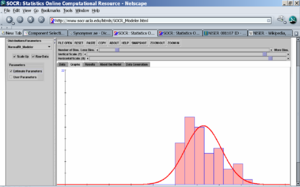
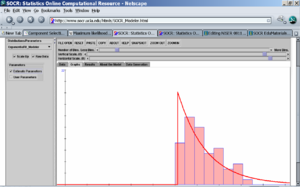
- We can easily compute the histogram of the average (per lake) mercury measurements and fit in Normal, Exponential or other distribution models to the histogram of the average mercury measurement using SOCR Charts and SOCR Modeler (see examples here). The left image below shows the fit of a \(Normal (\mu=10.54, \sigma^2=45.64)\) distribution model and the right image shows the (offset by 0.8) \(Exponential(\lambda = 10.54)\) model fit to the mercury histogram. The parameters in both cases are automatically calculated from the data using maximum likelihood estimation.
- We can also demonstrate the effect of using a wavelet decomposition of the column data (avg_mercury). The image below demonstrates the wavelet representation of this column vector data using Daubechies' 2 wavelet basis, where only the largest 10% of the wavelet coefficients were used to reconstruct/synthesize the (compressed) wavelet representation of the data. More information about wavelets and wavelets-based activities may be found here. You can use the SOCR Modeler to fit is any of a number of wavelet models to data and observe the effect of wavelet shrinkage on the corresponding wavelet model fit.
- We can easily compute the histogram of the average (per lake) mercury measurements and fit in Normal, Exponential or other distribution models to the histogram of the average mercury measurement using SOCR Charts and SOCR Modeler (see examples here). The left image below shows the fit of a \(Normal (\mu=10.54, \sigma^2=45.64)\) distribution model and the right image shows the (offset by 0.8) \(Exponential(\lambda = 10.54)\) model fit to the mercury histogram. The parameters in both cases are automatically calculated from the data using maximum likelihood estimation.
Statistics
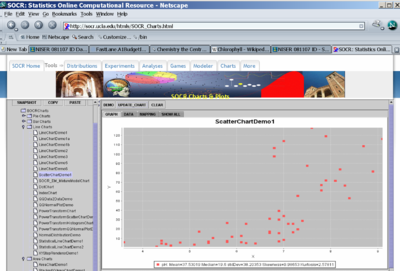
- Are there associations in these data between Alkalinity, pH, Calcium and Chlorophyll? Using SOCR Analyses construct regression plots illustrating the relations between Calcium, Chlorophyll and Avg_Mercury, as shown on the image below.
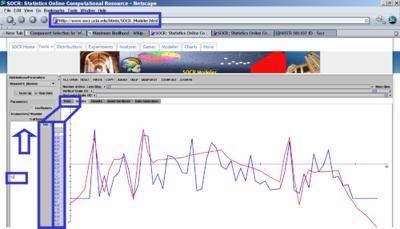
- Generate some exploratory data analyses (EDA plots). For instance, the figure below demonstrates how you can use SOCR Charts to obtain the scatter plot of Alkalinity vs. pH. What are some inferences about the bivariate relations between these variables?
- What is the distribution of the average amount of mercury in the entire sample? We can use SOCR Charts to construct the histogram of the avg_mercury column data. To learn how to use SOCR Charts see these Charts Activities. Notice the effect the bin-size parameter has on the shape of the data histogram (you can smoothly vary this parameter). Although, there are many choices for determining the bin-size parameter, in general, the optimal choice of the histogram bin-size parameter may be determined by \({2\times(IQR)} \over {N^{1\over 3}}\), where the interquartile range \(IQR= Q_3 - Q_1\), and \(Q_3\) and \(Q_1\) are the third and first quartiles for the sample data, respectively.
- Is there evidence of statistical differences between the two groups (according to the age_data variable) in either Alkalinity or pH?
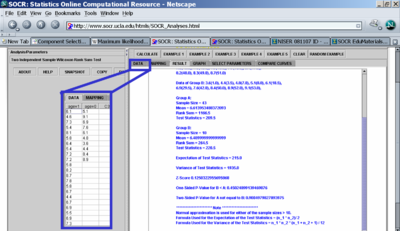
- There does not seem to be a statistically significant difference in the pH levels between the two groups separated by the age_data varaible. A non-parametric Wilcoxon Rank Sum test did not produce significantly small p-value (see the image below showing the result of the calculations using SOCR Analyses.
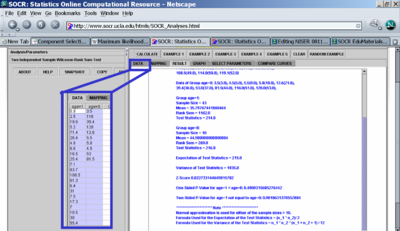
- Similarly, there does not seem to be a statistically significant difference in the Alkalinity levels between the two groups separated by the age_data variable. The same non-parametric Wilcoxon Rank Sum test did not produce significantly small p-value (see the image below showing the result of the calculations using SOCR Analyses.
- There does not seem to be a statistically significant difference in the pH levels between the two groups separated by the age_data varaible. A non-parametric Wilcoxon Rank Sum test did not produce significantly small p-value (see the image below showing the result of the calculations using SOCR Analyses.
Computational Resources
- Internet-based SOCR Tools (including offline resources, e.g., tables)
Hands-on activities
- TBD (Step-by-step practice problems).
Notes
- Follow the directions in the SOCR Wiki Editing Guide to expand, revise or improve these materials.
- NISER Home page: http://www.NSER.org
"-----
Translate this page: