Difference between revisions of "SMHS ModelFitting"
(→Scientific Methods for Health Sciences - Model Fitting and Model Quality (KS-test)) |
|||
| Line 48: | Line 48: | ||
plot(ecdf(control),verticals=TRUE,main='Cumulative Fraction Plot') | plot(ecdf(control),verticals=TRUE,main='Cumulative Fraction Plot') | ||
| + | [[Image:SMHS_Fig_1_Model_Fitting.png|500px]] | ||
| + | *Now plot the control group using a log scale, which will give more space to display the small x data points. Now the plot seems to present the data points evenly into two halves (half above the median, half below the median), which is a little bit below 1. | ||
| + | [[Image:SMHS_Fig_2_Model_Fitting.png|500px]] | ||
| + | log.con <- log(control) | ||
| + | plot(ecdf(log.con),verticals=T, main='Cumulative Fraction Plot of Log(Control)') | ||
| + | *Now, plot the cumulative fraction of both the treatment and the control group on the same graph. | ||
| + | [[Image:SMHS_Fig_3_Model_Fitting.png|500px]] | ||
| + | From the chart, we can see that both datasets span much of the same range of the values, but for most of the x value, the fraction of the treatment (red) is less than the fraction of the control group (blue). We denote the difference in the two fractions at each x value and the K-S test uses the maximum vertical deviation between the two curves. In this case, the maximum deviation occurs near x=1 and D=0.45. The fraction of the treatment group that is less than 1 is 0.2 (4 out of 20), and that for the control group is 0.65 (13 out of the 20 values), thus the maximum difference in the cumulative fraction is D=0.45). | ||
| + | Note that: the value of D is not affected by scale changes like using log, which is different from the t-statistics. Hence, the K-S test is a robust test that cares only about the relative distribution of the data. | ||
| + | log.treat <- log(treatment) | ||
| + | plot(ecdf(log.con),verticals=TRUE, main='Cumulative Fraction Plot on Log | ||
| + | Scale',col.p='bisque',col.h='blue',col.v='blue',xlim=c(-3,5)) | ||
| + | par(new=T) | ||
| + | plot(ecdf(log.treat),verticals=TRUE, | ||
| + | col.p='bisque',col.h='red',col.v='red',main='',xlim=c (-3,5)) | ||
| + | con.p <- ecdf(log.con) | ||
| + | treat.p <- ecdf(log.treat) | ||
| + | con.p(0)-treat.p(0) ### D=0.45 | ||
| + | (ecdf(control))(1)-(ecdf(treatment))(1) ## D=0.45 same as using the log-scale | ||
| + | '''4) Using the Percentile Plot:''' for our habit of observing and comparing continuous curves. We may seek to use something similar to cumulative fraction plot, but without the odd steps, say, try the percentiles. Consider the dataset of ${-0.45, 1.11, 0.48, -0.82, -1.26}.$ Sort this data from small to large ${-1.26, -0.82, -0.45, 0.48, 1.11}.$ The median is $-0.45$, which is the 50th percentile. To calculate the percentile, denote the point’s location in the sorted dataset as r, and then divided by the number of points plus one: percentile = $r/(N+1)$. Now we have the set of (data, percentile) pairs: ${(-1.26, 0.167), (-0.82, 0.333), (-0.45, 0.5), (0.48, 0.667), (1.11, 0.833)}$. We can connect the adjacent data points with a straight line and the resulting collection of connected straight line segment is called an ''ogive'''''Bold text'''). | ||
| + | RCODE: | ||
| + | data <- c(-0.45, 1.11, 0.48, -0.82, -1.26) | ||
| + | sort.data <- sort(data) | ||
| + | percentile <- c(0.167, 0.333, 0.5, 0.667, 0.8333) | ||
| + | plot(ecdf(data),verticals=T,xlim=c(-1.5,1.5),ylim=c(0,1), xlab='Data', ylab='' ,main='Cumulative Fraction Plot vs. Percentil Plot') | ||
| + | par(new=T) | ||
| + | plot(sort.data,percentile,type='o',xlim=c(-1.5,1.5),ylim=c(0,1),col=2,xlab='', ylab='', main='') | ||
| + | [[Image:SMHS_Fig_4_Model_Fitting.png|500px]] | ||
| + | Reasons to prefer percentile plot to cumulative fraction plots: the percentile plot is a better estimate of the distribution function and the percentiles allow us to use ‘probability graph paper’, plots with specially scaled axis divisions. Probability scales on the y-axis allow us to see how normal the data is. Normally distributed data will show a straight line on the probability paper while log-normal data will show a straight line with probability-log scaled axes. | ||
| − | + | '''5) The K-S statistic in more than one dimension:''' modifies the K-S test statistic to accommodate the multivariate data. Given that the maximum difference between two joint $cdf$ is not generally the same as the maximum difference of any of the complementary distribution functions, the modification is not straightforward in this way. Instead, the maximum difference will differ depending on which of $Pr(x<X \Lambda Y>y)$ or any of the other two possible arrangement is used. One approach may be to compare the cdfs of the two samples with all possible orderings, and take the largest as the K-S statistic. In $d$ dimensions, there are $2^{d}-1$ orderings. The critical values for the test statistic can be obtained by simulations, but depend on the dependence structure in the joint distribution. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Applications=== | ===Applications=== | ||
| Line 146: | Line 112: | ||
===Problems=== | ===Problems=== | ||
| − | + | 1) Consider the following dataset, control={0.22, -0.87, -2.39, -1.79, 0.37, -1.54, 1.28, -0.31, -0.74, 1.72, 0.38, -0.17, -0.62, -1.10, 0.30, 0.15, 2.30, 0.19, -0.50, -0.09}, treatment={-5.13, -2.19, -2.43, -3.83, 0.50, -3.25, 4.32, 1.63, 5.18, -0.43, 7.11, 4.87, -3.10, -5.81, 3.76, 6.31, 2.58, 0.07, 5.76, 3.50}, use the K-S test to test the equality of the two samples. Include all necessary steps and plots. | |
| − | + | 2) Revise the example given in the lecture with a different control group: control={1.26, 0.34, 0.70, 1.75, 50.57, 1.55, 0.08, 0.42, 0.50, 3.20, 0.15, 0.49, 0.95, 0.24, 1.37, 0.17, 6.98, 0.10, 0.94, 0.38}, run the K-S test again and state clearly your conclusions with necessary plots. | |
| − | + | 3) Two types of plants are in bloom in wood and we want to study if bees prefer one tree to the other? We collect data by using a stop-watch to time how long a bee stays near a particular tree. We begin to time when the bee touches the tree; we stop timing when the bee is more than a meter from the tree. (As a result all our times are at least 1 second long: it takes a touch-and-go bee that long to get one meter from the tree.) We wanted to time exactly the same number of bees for each tree, but it started to rain. (Unequal dataset size is not a problem for the KS-test.) Apply the K-S test with this example. The data are given as below: | |
T1={23.4, 30.9, 18.8, 23.0, 21.4, 1, 24.6, 23.8, 24.1, 18.7, 16.3, 20.3, 14.9, 35.4, 21.6, 21.2, 21.0, 15.0, 15.6, 24.0, 34.6, 40.9, 30.7, 24.5, 16.6, 1, 21.7, 1, 23.6, 1, 25.7, 19.3, 46.9, 23.3, 21.8, 33.3, 24.9, 24.4, 1, 19.8, 17.2, 21.5, 25.5, 23.3, 18.6, 22.0, 29.8, 33.3, 1, 21.3, 18.6, 26.8, 19.4, 21.1, 21.2, 20.5, 19.8, 26.3, 39.3, 21.4, 22.6, 1, 35.3, 7.0, 19.3, 21.3, 10.1, 20.2, 1, 36.2, 16.7, 21.1, 39.1, 19.9, 32.1, 23.1, 21.8, 30.4, 19.62, 15.5} | T1={23.4, 30.9, 18.8, 23.0, 21.4, 1, 24.6, 23.8, 24.1, 18.7, 16.3, 20.3, 14.9, 35.4, 21.6, 21.2, 21.0, 15.0, 15.6, 24.0, 34.6, 40.9, 30.7, 24.5, 16.6, 1, 21.7, 1, 23.6, 1, 25.7, 19.3, 46.9, 23.3, 21.8, 33.3, 24.9, 24.4, 1, 19.8, 17.2, 21.5, 25.5, 23.3, 18.6, 22.0, 29.8, 33.3, 1, 21.3, 18.6, 26.8, 19.4, 21.1, 21.2, 20.5, 19.8, 26.3, 39.3, 21.4, 22.6, 1, 35.3, 7.0, 19.3, 21.3, 10.1, 20.2, 1, 36.2, 16.7, 21.1, 39.1, 19.9, 32.1, 23.1, 21.8, 30.4, 19.62, 15.5} | ||
T2={16.5, 1, 22.6, 25.3, 23.7, 1, 23.3, 23.9, 16.2, 23.0, 21.6, 10.8, 12.2, 23.6, 10.1, 24.4, 16.4, 11.7, 17.7, 34.3, 24.3, 18.7, 27.5, 25.8, 22.5, 14.2, 21.7, 1, 31.2, 13.8, 29.7, 23.1, 26.1, 25.1, 23.4, 21.7, 24.4, 13.2, 22.1, 26.7, 22.7, 1, 18.2, 28.7, 29.1, 27.4, 22.3, 13.2, 22.5, 25.0, 1, 6.6, 23.7, 23.5, 17.3, 24.6, 27.8, 29.7, 25.3, 19.9, 18.2, 26.2, 20.4, 23.3, 26.7, 26.0, 1, 25.1, 33.1, 35.0, 25.3, 23.6, 23.2, 20.2, 24.7, 22.6, 39.1, 26.5, 22.7} | T2={16.5, 1, 22.6, 25.3, 23.7, 1, 23.3, 23.9, 16.2, 23.0, 21.6, 10.8, 12.2, 23.6, 10.1, 24.4, 16.4, 11.7, 17.7, 34.3, 24.3, 18.7, 27.5, 25.8, 22.5, 14.2, 21.7, 1, 31.2, 13.8, 29.7, 23.1, 26.1, 25.1, 23.4, 21.7, 24.4, 13.2, 22.1, 26.7, 22.7, 1, 18.2, 28.7, 29.1, 27.4, 22.3, 13.2, 22.5, 25.0, 1, 6.6, 23.7, 23.5, 17.3, 24.6, 27.8, 29.7, 25.3, 19.9, 18.2, 26.2, 20.4, 23.3, 26.7, 26.0, 1, 25.1, 33.1, 35.0, 25.3, 23.6, 23.2, 20.2, 24.7, 22.6, 39.1, 26.5, 22.7} | ||
Revision as of 08:55, 21 October 2014
Contents
[hide]Scientific Methods for Health Sciences - Model Fitting and Model Quality (KS-test)
Overview
Kolmogorov-Smirnov Test (K-S test) is a nonparametric test commonly applied in various fields to test on the equality of continuous, one-dimensional probability distribution that can be used to compare one sample with a reference probability distribution, which is commonly referred to one-sample K-S test, or to compare two samples, which is referred to as two-sample K-S test. The K-S test is used to determine if two datasets differ significantly. In this lecture, we are going to present a general introduction to K-S test and illustrate its application with examples. The implementation of K-S test in the statistical package R will also be discussed with specific examples.
Motivation
When we talk about testing the equality of two dataset, the first idea came into our mind may always be the simple t-test. However, there are situations where it is a mistake to trust the result of a t-test. Consider the situation where the control and treatment groups do not differ in mean, but only in some other way. Consider two dataset with same mean and very different variations, in this situation, the t-test cannot see the difference. Situations where the treatment and control groups are smallish datasets (say less than 20) that differ in mean but substantial non-normal distribution masks the difference. Consider two datasets drawn from lognormal distributions that differ substantially in mean, in this situation, the t-test also fails. Among all those situations, K-S test would be the answer. How does the K-S test work?
Theory
1) K-S test: a nonparametric test commonly applied in various fields to test on the equality of continuous, one-dimensional probability distribution that can be used to compare one sample with a reference probability distribution or to compare two samples. The K-S test quantifies a distance between the empirical distribution function of the sample and the cdf of reference distribution or between the empirical distribution functions of two samples. The null hypothesis is that the samples are drawn from the same distribution or that the sample is drawn from the reference distribution.
- K-S test is sensitive to difference both in location and shape of the empirical cdf of the samples and is widely used as the nonparametric methods for comparing two samples.
- The empirical distribution function $F_{n}$ for n i.i.d. observations $X_{i}: F_{n} (x)=1/n \sum_{i=1}^{n} I_{X_{i} \leq x},$ where $I_{X_{i} \leq x}$ is the indicator function, which equals to 1 when $X_{i} \leq x$ and equals to $0$ otherwise.
- The K-S statistic for a given $cdf F(x)$ is $D_{n}=sup_{x}|F_{n}(x)-F(x)|,$ where sup{x} is the supremum of the set of distances. By the Gilvenko-Cantelli therorem, if the sample comes from distribution $F(x)$, then $D_{n}$ converges to $0$ almost surely in the limit when $n$ goes to infinity.
- Kolmogorov distribution (the distribution of the random variable): $K=sup_{t\in [0,1]} |B(t)|,$ where $B(t)$ is the Brownian bridge. The $c.d.f$ of $K$ is given: $Pr(K \leq x)=1-2 \sum_{k=1}^{\infty}(-1)^{k-1}e^{-2k^{2} x^{2}}=\frac {\sqrt{2 \pi}}{x} \sum_{k=1}^{\infty}e^{-(2k-1)^{2} \pi^{2} \setminus (8x^{2})}.$ Under the null hypothesis, the sample comes from the hypothesized distribution $F(x), \sqrt{n} D_{n} \overset{n\rightarrow \infty}{\rightarrow} sup_{t} |B(F(t))|$ in distribution, where $B(t)$ is the Brownian bridge. When $F$ is continuous, $\sqrt{n} D_{n}$ under the null hypothesis converges to the Kolmogorov distribution, which does not depend on $F.$
- The goodness-of-fit test or the K-S test is constructed by using the critical values of the Kolmogorov distribution. The null hypothesis is rejected at level $\alpha$ if $\sqrt{n} D_{n} > K_{\alpha},$ where $K_{\alpha}$ is found from $Pr(K \leq K_{\alpha})=1- \alpha.$ The asymptotic power of this test is $1$.
- Test with estimated parameters: if either the form or the parameters of $F(x)$ are determined from the data $X_{j}$ the critical values determined in this way are invalid. Modifications required to the test statistics and critical values have been proposed.
2) Two-sample K-S test: to test whether two underlying one-dimensional probability distribution differ. The K-S statistic is $D_{n,{n}'}=sup_{x} |F_{1,n}(x)-F_{2,{n}'}(x)|$, where $F_{1,n}$ and $F_{2,{n}'}$ are the empirical distribution functions of the first and second sample respectively. The null hypothesis is rejected at least $\alpha$ if $D_{n,{n}'}>c(\alpha)\sqrt{\frac{n+{n}'}{n{n}'}}$. The value of $c(\alpha)$ is given in the following table for each level of $\alpha.$
| $\alpha$ | 0.1 | 0.05 | 0.025 | 0.01 | 0.005 | 0.001 |
| c($\alpha$) | 1.22 | 1.36 | 1.48 | 1.63 | 1.73 | 1.95 |
Note: two-sample test checks whether the two data samples come from the same distribution. This does not specify what that common distribution is. While the K-S test is usually used to test whether a given $F(x)$ is the underlying probability distribution $F_{n}(x)$, the procedure may be inverted to give confidence limits on $F(x)$ itself. If we choose a critical value of the test statistics $D_{\alpha}$ such that $P(D_{n}>D_{\alpha})=\alpha$, then a band of width $\pm D_{\alpha}$ around $F_{n}(x)$ will entirely contain $F(x)$ with probability $1-\alpha.$
3) Illustration on how the K-S test works with example: consider the two datasets control$={1.26, 0.34, 0.70, 1.75, 50.57, 1.55, 0.08, 0.42, 0.50, 3.20, 0.15, 0.49, 0.95, 0.24, 1.37, 0.17, 6.98, 0.10, 0.94, 0.38}$, treatment= ${2.37, 2.16, 14.82, 1.73, 41.04, 0.23, 1.32, 2.91, 39.41, 0.11, 27.44, 4.51, 0.51, 4.50, 0.18, 14.68, 4.66, 1.30, 2.06, 1.19}$.
- Descriptive statistics: to reduce the list of all the data items to a few simpler numbers. For the control group, the mean is 3.607, the median is 0.60, the highest is 50.57, the lowest is 0.08 and the standard deviation is 11.165. Obviously, this dataset is not normally distributed.
- Sort the control data: control sorted$={0.08, 0.10, 0.15, 0.17, 0.24, 0.34, 0.38, 0.42, 0.49, 0.50, 0.70, 0.94, 0.95, 1.26, 1.37, 1.55, 1.75, 3.20, 6.98, 50.57},$ evidently no data lies strictly below $0.08, 5% = 1/20$ of the data is smaller than $0.1$, 10% of the data is strictly smaller than $0.15, 15%$ of the data is strictly smaller than $0.17$, $\cdots.$ There are $17$ data points smaller than $\pi$, and we say that the cumulative fraction of the data smaller than $\pi$ is $17/20 (0.85).$
RCODE: Control <- c(1.26, 0.34, 0.70, 1.75, 50.57, 1.55, 0.08, 0.42, 0.50, 3.20, 0.15, 0.49, 0.95 , 0.24, 1.37, 0.17, 6.98, 0.10, 0.94, 0.38) Treatment <- c(2.37, 2.16, 14.82, 1.73, 41.04, 0.23, 1.32, 2.91, 39.41, 0.11, 27.44, 4.51, 0.51, 4.50, 0.18, 14.68, 4.66, 1.30, 2.06, 1.19) summary(control) Min. 1st Qu. Median Mean 3rd Qu. Max. 0.080 0.315 0.600 3.607 1.415 50.570 sd(control) [1] 11.16464
- Plot the empirical cumulative plot of the control data. From the chart, we can see that the majority of the data fall in the a small fraction of the plot on the top left, so this is a clear sign that the dataset from control group does not follow a normal distribution.
library(stats) ecdf(control) plot(ecdf(control),verticals=TRUE,main='Cumulative Fraction Plot')
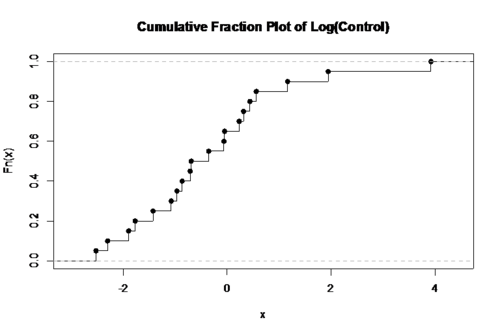
- Now plot the control group using a log scale, which will give more space to display the small x data points. Now the plot seems to present the data points evenly into two halves (half above the median, half below the median), which is a little bit below 1.
log.con <- log(control) plot(ecdf(log.con),verticals=T, main='Cumulative Fraction Plot of Log(Control)')
- Now, plot the cumulative fraction of both the treatment and the control group on the same graph.
From the chart, we can see that both datasets span much of the same range of the values, but for most of the x value, the fraction of the treatment (red) is less than the fraction of the control group (blue). We denote the difference in the two fractions at each x value and the K-S test uses the maximum vertical deviation between the two curves. In this case, the maximum deviation occurs near x=1 and D=0.45. The fraction of the treatment group that is less than 1 is 0.2 (4 out of 20), and that for the control group is 0.65 (13 out of the 20 values), thus the maximum difference in the cumulative fraction is D=0.45). Note that: the value of D is not affected by scale changes like using log, which is different from the t-statistics. Hence, the K-S test is a robust test that cares only about the relative distribution of the data.
log.treat <- log(treatment) plot(ecdf(log.con),verticals=TRUE, main='Cumulative Fraction Plot on Log Scale',col.p='bisque',col.h='blue',col.v='blue',xlim=c(-3,5)) par(new=T) plot(ecdf(log.treat),verticals=TRUE, col.p='bisque',col.h='red',col.v='red',main=,xlim=c (-3,5)) con.p <- ecdf(log.con) treat.p <- ecdf(log.treat) con.p(0)-treat.p(0) ### D=0.45 (ecdf(control))(1)-(ecdf(treatment))(1) ## D=0.45 same as using the log-scale
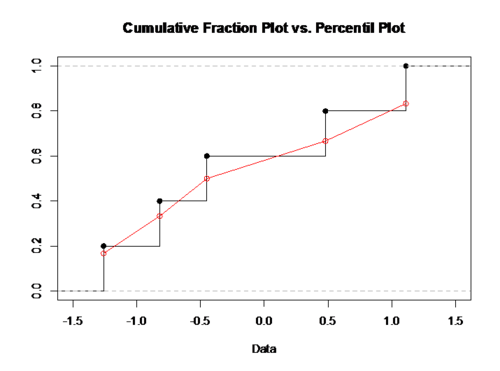
4) Using the Percentile Plot: for our habit of observing and comparing continuous curves. We may seek to use something similar to cumulative fraction plot, but without the odd steps, say, try the percentiles. Consider the dataset of ${-0.45, 1.11, 0.48, -0.82, -1.26}.$ Sort this data from small to large ${-1.26, -0.82, -0.45, 0.48, 1.11}.$ The median is $-0.45$, which is the 50th percentile. To calculate the percentile, denote the point’s location in the sorted dataset as r, and then divided by the number of points plus one: percentile = $r/(N+1)$. Now we have the set of (data, percentile) pairs: ${(-1.26, 0.167), (-0.82, 0.333), (-0.45, 0.5), (0.48, 0.667), (1.11, 0.833)}$. We can connect the adjacent data points with a straight line and the resulting collection of connected straight line segment is called an ogiveBold text).
RCODE: data <- c(-0.45, 1.11, 0.48, -0.82, -1.26) sort.data <- sort(data) percentile <- c(0.167, 0.333, 0.5, 0.667, 0.8333) plot(ecdf(data),verticals=T,xlim=c(-1.5,1.5),ylim=c(0,1), xlab='Data', ylab= ,main='Cumulative Fraction Plot vs. Percentil Plot') par(new=T) plot(sort.data,percentile,type='o',xlim=c(-1.5,1.5),ylim=c(0,1),col=2,xlab=, ylab=, main=)
Reasons to prefer percentile plot to cumulative fraction plots: the percentile plot is a better estimate of the distribution function and the percentiles allow us to use ‘probability graph paper’, plots with specially scaled axis divisions. Probability scales on the y-axis allow us to see how normal the data is. Normally distributed data will show a straight line on the probability paper while log-normal data will show a straight line with probability-log scaled axes.
5) The K-S statistic in more than one dimension: modifies the K-S test statistic to accommodate the multivariate data. Given that the maximum difference between two joint $cdf$ is not generally the same as the maximum difference of any of the complementary distribution functions, the modification is not straightforward in this way. Instead, the maximum difference will differ depending on which of $Pr(x<X \Lambda Y>y)$ or any of the other two possible arrangement is used. One approach may be to compare the cdfs of the two samples with all possible orderings, and take the largest as the K-S statistic. In $d$ dimensions, there are $2^{d}-1$ orderings. The critical values for the test statistic can be obtained by simulations, but depend on the dependence structure in the joint distribution.
Applications
1)This article presents the SOCR analyses example on Kolmogorov-Smirnoff Test, which compares how distinct two values are. It presents a general introduction to the K-S test and illustrates its application with the Oats example and control-treatment test through the SOCR program.
2) This article presents the SOCR activity which demonstrate the random sampling and fitting of mixture models to data. It starts with a general introduction to the mixture-model distribution and data description, followed with the exploratory data analysis and model fitting in the SOCR environment.
3) This article presented on the Kolmogorov-Smirnov statistical test for the analysis of histograms and discussed about the test for both the two-sample case (comparing fn1(X) to fn2 (X)) and the one-sample case (comparing fn1 (X) to f(X)). Presentation of the specific algorithmic steps involved is done through development of an example where the data are from an experiment discussed elsewhere in this issue. It is shown that the two histograms examined come from two different parent populations at the 99.9% confidence level.
4) This article developed the digital techniques for detecting changes between two Landsat images. A data matrix containing 16 Ã 16 picture elements was used for this purpose. The Landsat imagery data was corrected for sensor inconsistencies and varying sun illumination. The Kolmogorov-Smirnov test (K-S test) was performed between the two corrected data matrices. This test is based on the absolute value of the maximum difference (Dmax) between the two cumulative frequency distributions. The limiting distribution of Dmax is known; thus a statistical decision concerning changes can be evaluated for the region. The K-S test was applied to different test sites. It successfully isolated regions of change. The test was found to be relatively independent of slight misregistration.
Software
http://stat.ethz.ch/R-manual/R-patched/library/stats/html/ks.test.html
http://astrostatistics.psu.edu/su07/R/html/stats/html/ks.test.html
RCODE are attached as in the examples given in this lecture.
Problems
1) Consider the following dataset, control={0.22, -0.87, -2.39, -1.79, 0.37, -1.54, 1.28, -0.31, -0.74, 1.72, 0.38, -0.17, -0.62, -1.10, 0.30, 0.15, 2.30, 0.19, -0.50, -0.09}, treatment={-5.13, -2.19, -2.43, -3.83, 0.50, -3.25, 4.32, 1.63, 5.18, -0.43, 7.11, 4.87, -3.10, -5.81, 3.76, 6.31, 2.58, 0.07, 5.76, 3.50}, use the K-S test to test the equality of the two samples. Include all necessary steps and plots.
2) Revise the example given in the lecture with a different control group: control={1.26, 0.34, 0.70, 1.75, 50.57, 1.55, 0.08, 0.42, 0.50, 3.20, 0.15, 0.49, 0.95, 0.24, 1.37, 0.17, 6.98, 0.10, 0.94, 0.38}, run the K-S test again and state clearly your conclusions with necessary plots.
3) Two types of plants are in bloom in wood and we want to study if bees prefer one tree to the other? We collect data by using a stop-watch to time how long a bee stays near a particular tree. We begin to time when the bee touches the tree; we stop timing when the bee is more than a meter from the tree. (As a result all our times are at least 1 second long: it takes a touch-and-go bee that long to get one meter from the tree.) We wanted to time exactly the same number of bees for each tree, but it started to rain. (Unequal dataset size is not a problem for the KS-test.) Apply the K-S test with this example. The data are given as below: T1={23.4, 30.9, 18.8, 23.0, 21.4, 1, 24.6, 23.8, 24.1, 18.7, 16.3, 20.3, 14.9, 35.4, 21.6, 21.2, 21.0, 15.0, 15.6, 24.0, 34.6, 40.9, 30.7, 24.5, 16.6, 1, 21.7, 1, 23.6, 1, 25.7, 19.3, 46.9, 23.3, 21.8, 33.3, 24.9, 24.4, 1, 19.8, 17.2, 21.5, 25.5, 23.3, 18.6, 22.0, 29.8, 33.3, 1, 21.3, 18.6, 26.8, 19.4, 21.1, 21.2, 20.5, 19.8, 26.3, 39.3, 21.4, 22.6, 1, 35.3, 7.0, 19.3, 21.3, 10.1, 20.2, 1, 36.2, 16.7, 21.1, 39.1, 19.9, 32.1, 23.1, 21.8, 30.4, 19.62, 15.5} T2={16.5, 1, 22.6, 25.3, 23.7, 1, 23.3, 23.9, 16.2, 23.0, 21.6, 10.8, 12.2, 23.6, 10.1, 24.4, 16.4, 11.7, 17.7, 34.3, 24.3, 18.7, 27.5, 25.8, 22.5, 14.2, 21.7, 1, 31.2, 13.8, 29.7, 23.1, 26.1, 25.1, 23.4, 21.7, 24.4, 13.2, 22.1, 26.7, 22.7, 1, 18.2, 28.7, 29.1, 27.4, 22.3, 13.2, 22.5, 25.0, 1, 6.6, 23.7, 23.5, 17.3, 24.6, 27.8, 29.7, 25.3, 19.9, 18.2, 26.2, 20.4, 23.3, 26.7, 26.0, 1, 25.1, 33.1, 35.0, 25.3, 23.6, 23.2, 20.2, 24.7, 22.6, 39.1, 26.5, 22.7} Note that this example is based on data distributed according to the Cauchy distribution: a particularly abnormal case and the plots do not look particularly abnormal, however the large number of outliers is a tip off of a non-normal distribution.
References
- SOCR Home page: http://www.socr.umich.edu
Translate this page: