SMHS BigDataBigSci SEM
Model-based Analytics - Structural Equation Modeling (SEM)
Questions
• How to represent dependencies in linear models and examine causal effects?
• Is there a way to study population average effects of a covariate against specific individual effects?
Overview
SEM allow re-parameterization of random-effects to specify latent variables that may affect measures at different time points using structural equations. SEM show variables having predictive (possibly causal) effects on other variables (denoted by arrows) where coefficients index the strength and direction of predictive relations. SEM does not offer much more than what classical regression methods do, but it does allow simultaneous estimation of multiple equations modeling complementary relations.
GEE is a marginal longitudinal method that directly assesses the mean relations of interest (i.e., how the mean dependent variable changes over time), accounting for covariances among the observations within subjects, and getting a better estimate and valid significance tests of the relations. Thus, GEE estimates two different equations, (1) for the mean relations, and (2) for the covariance structure. An advantage of GEE over random-effect models is that it does not require the dependent variable to be normally distributed. However, a disadvantage of GEE is that it is less flexible and versatile – commonly employed algorithms for it require a small-to-moderate number of time points evenly (or approximately evenly) spaced, and similarly spaced across subjects. Nevertheless, it is a little more flexible than repeated-measure ANOVA because it permits some missing values and has an easy way to test for and model away the specific form of autocorrelation within subjects.
GEE is mostly used when the study is focused on uncovering the population average effect of a covariate vs. the individual specific effect. These two things are only equivalent for linear models, but not in non-linear models.
FIX FORMULA For instance, suppose Yi, j, is the random effects logistic model of the jth, observation of the ith subject, then log(p_(i,j)/(1-p_(i,j) ))=μ+ν_i, where ν_i~N(0,σ^2) is a random effect for subject i and p_(i,j)=P(Y_(i,j)=1|ν_i).
(1) When using a random effects model on such data, the estimate of μ accounts for the fact that a mean zero normally distributed perturbation was applied to each individual, making it individual-specific.
(2) When using a GEE model on the same data, we estimate the population average log odds,
FIX FORMULA δ=log((E_ν (1/(1+e^(-μ+ν_i ) )))/(1-E_ν (1/(1+e^(-μ+ν_i ) )) )), in general μ≠δ. If μ=1 and σ^2=1, then δ≈.83.
empirically:
m <- 1; s <- 1; v<-rnorm(1000, 0,s); v2 <- 1/(1+exp(-m+v)); v_mean <- mean(v2)
d <- log(v_mean/(1-v_mean)); d
Note that the random effects have mean zero on the transformed, linked, scale, but their effect is not mean zero on the original scale of the data. We can also simulate data from a mixed effects logistic regression model and compare the population level average with the inverse-logit of the intercept to see that they are not equal. This leads to a difference of the interpretation of the coefficients between GEE and random effects models, or SEM.
That is, there will be a difference between the GEE population average coefficients and the individual specific coefficients (random effects models).
# theoretically, if it can be computed:
FIX FORMULAS!! E(Y)=μ=1 (in this specific case), but the expectation of the population average log odds δ= log[(P(Y_(i,j)=1|ν_i))/(1-P(Y_(i,j)=1|ν_i))] would be < 1 . Note that this is kind of related to the fact that a grand-total average need not be equal to an average of partial averages.
The mean of the ith person in the jth observation (e.g., location, time, etc.) can be expressed by: E(Yij | Xij,α_j)= g[μ(Xij ┤|β)+Uij(α_j,Xij)], Where μ(X_ij |β) is the average “response” of a person with the same covariates X_ij, β a set of fixed effect coefficients, and Uij(α_j,Xij) is an error term that is a function of the (time, space) random effects, α_j, and also a function of the covariates X_ij, and g is the link function which specifies the regression type -- e.g., linear: g^(-1) (u)=u, log: g^(-1) (u)= log(u), logistic: g^(-1) (u)=log(u/(1-u)) E(Uij(α_j,Xij)|Xij)=0.
The link function, g(u), provides the relationship between the linear predictor and the mean of the distribution function. For practical applications there are many commonly used link functions. It makes sense to try to match the domain of the link function to the range of the distribution function's mean.
INSERT TABLE!!!!!!!!!!!
Structural Equation Modeling (SEM)
SEM is a general multivariate statistical analysis technique that can be used for causal modeling/inference, path analysis, confirmatory factor analysis (CFA), covariance structure modeling, and correlation structure modeling.
Advantages
• It allows testing models with multiple dependent variables
• Provides mechanisms for modeling mediating variables
• Enables modeling of error terms
• Facilitates modeling of challenging data (longitudinal with auto-correlated errors, multi-level data, non-normal data, incomplete data)
This method SEM allows separation of observed and latent variables. Other standard statistical procedures may be viewed as special cases of SEM, where statistical significance less important, than in other techniques, and covariances are the core of structural equation models.
Definitions
- The disturbance, D, is the variance in Y unexplained by a variable X that is assumed to affect Y.
X → Y ← D
- Measurement error, E, is the variance in X unexplained by A, where X is an observed variable that is presumed to measure a latent variable, A.
A → X ← E
- Categorical variables in a model are exogenous (independent) or endogenous (dependent).
Notation
- In SEM observed (or manifest) indicators are represented by squares/rectangles whereas latent variables (or factors) represented by circles/ovals.
PLEASE FIX ARROWS *Relations: Direct effects (→), Reciprocal effects (<--> or ), and Correlation or covariance ( ) all have different appearance in SEM models.
Model Components
The measurement part of SEM model deals with the latent variables and their indicators. A pure measurement model is a confirmatory factor analysis (CFA) model with unmeasured covariance (bidirectional arrows) between each possible pair of latent variables. There are straight arrows from the latent variables to their respective indicators and straight arrows from the error and disturbance terms to their respective variables, but no direct effects (straight arrows) connecting the latent variables. The measurement model is evaluated using goodness of fit measures (Chi-Square test, BIC, AIC, etc.) Validation of the measurement model is always first.
Then we proceed to the structural model (including a set of exogenous and endogenous variables together with the direct effects (straight arrows) connecting them along with the disturbance and error terms for these variables that reflect the effects of unmeasured variables not in the model).
Notes
• Sample-size considerations: mostly same as for regression - more is always better
• Model assessment strategies: Chi-square test, Comparative Fit Index, Root Mean Square Error, Tucker Lewis Index, Goodness of Fit Index, AIC, and BIC.
• Choice for number of Indicator variables: depends on pilot data analyses, a priori concerns, fewer is better.
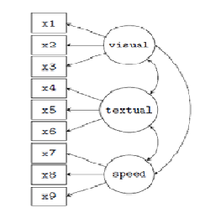
Hands-on Example 1 (School Kids Mental Abilities)
These data (Holzinger & Swineford 1939) include mental ability test scores of 7 & 8 grade children from two schools (Pasteur and Grant-White). This version of the dataset includes only 9 (out of the 26) tests. We can build and test a confirmatory factor analysis (CFA) SEM model for 3 correlated latent variables (or factors), each with three indicators:
o visual factor measured by 3 variables: x1, x2 and x3,
o textual factor measured by 3 variables: x4, x5 and x6,
o speed factor measured by 3 variables: x7, x8 and x9.
| ID | lhs | op | rhs | user | free | ustart |
| 1 | Visual | =~ | x1 | 1 | 0 | 1 |
| 2 | Visual | =~ | x2 | 1 | 1 | NA |
| 3 | Visual | =~ | x3 | 1 | 2 | NA |
| 4 | Textual | =~ | x4 | 1 | 0 | 1 |
| 5 | Textual | =~ | x5 | 1 | 3 | NA |
| 6 | Textual | =~ | x6 | 1 | 4 | NA |
| 7 | Speed | =~ | x7 | 1 | 0 | 1 |
| 8 | Speed | =~ | x8 | 1 | 5 | NA |
| 9 | Speed | =~ | x9 | 1 | 6 | NA |
| 10 | x1 | ~~ | x1 | 0 | 7 | NA |
| 11 | x2 | ~~ | x2 | 0 | 8 | NA |
| 12 | x3 | ~~ | x3 | 0 | 9 | NA |
| 13 | x4 | ~~ | x4 | 0 | 10 | NA |
| 14 | x5 | ~~ | x5 | 0 | 11 | NA |
| 15 | x6 | ~~ | x6 | 0 | 12 | NA |
| 16 | x7 | ~~ | x7 | 0 | 13 | NA |
| 17 | x8 | ~~ | x8 | 0 | 14 | NA |
| 18 | x9 | ~~ | x9 | 0 | 15 | 47.8 |
| 19 | Visual | ~~ | Visual | 0 | 16 | NA |
| 20 | Textual | ~~ | Textual | 0 | 17 | NA |
| 21 | Speed | ~~ | Speed | boy | 18 | NA |
| 22 | Visual | ~~ | Textual | girl | 19 | NA |
| 23 | Visual | ~~ | Speed | girl | 20 | NA |
| 24 | Textual | ~~ | Speed | boy | 21 | NA |
There are 3 latent variables (factors) in this model, each with 3 indicators, resulting in 9 factor loadings that need to be estimated. There are also 3 covariances among the latent variables {another three parameters}.
These 12 parameters are represented in the path diagram as single-headed and double-headed arrows, respectively. We also need to estimate the residual variances of the 9 observed variables and the variances of the 3 latent variables, resulting in 12 additional free parameters. In total, we have 24 parameters.
To fully identify the model we need to set the metric of the latent variables. There are 2 ways to do this:
o for each latent variable, fix the factor loading of one of the indicators (typically the first) to a constant (e.g., 1.0), or
o standardize the variances of the 3 latent variables.
Either way, we fix 3 of these 24 parameters, and 21 parameters remain free.
The parTable(fit) method, generates this table output.
The `rhs', `op' and `lhs' columns define the parameters of the model. All parameters with the `=~' operator are factor loadings, whereas all parameters with the `~~' operator are variances or covariances. Nonzero elements in the `free' column are the free parameters of the model. Zero elements in the `free' column correspond to fixed parameters, whose value is found in the `start' column.
Lavaan’s user-friendly model-specification approach is implemented in the fitting functions: cfa() and sem().
Since these data contain 3 latent variables, and no regressions, the minimalist syntax is:
data.model <- 'visual =~ x1 + x2 + x3 textual =~ x4 + x5 + x6 speed =~ x7 + x8 + x9'
Fit the CFA model:
fit.1 <- cfa(data.model, data = HolzingerSwineford1939)
The `user' column (parTabale) shows which parameters were explicitly contained in the user-specified model syntax (= 1), and which parameters were added by the cfa() function (= 0).
parTable(fit.1)
If we prefer not to fix the factor loadings of the first indicator, but instead want to fix the variances of the latent variances, the model syntax would be changed to:
fit.2 <- 'visual =~ NA*x1 + x2 + x3 textual =~ NA*x4 + x5 + x6 speed =~ NA*x7 + x8 + x9 visual ~~ 1*visual textual ~~ 1*textual speed ~~ 1*speed'
More complex model specifications can be made using the full lavaan model syntax:
fit.full <- ' # latent variables visual =~ 1*x1 + x2 + x3 textual =~ 1*x4 + x5 + x6 speed =~ 1*x7 + x8 + x9 # residual variances observed variables x1 ~~ x1 x2 ~~ x2 x3 ~~ x3 x4 ~~ x4 x5 ~~ x5 x6 ~~ x6 x7 ~~ x7 x8 ~~ x8 x9 ~~ x9 # factor variances visual ~~ visual textual ~~ textual speed ~~ speed # factor covariances visual ~~ textual + speed textual ~~ speed' fit.3 <- lavaan(fit.full, data = HolzingerSwineford1939)
We can specify the model where the first factor loadings are explicitly fixed to one, and the covariances among the factors are added manually.
fit.mixed <- ' # latent variables visual =~ 1*x1 + x2 + x3 textual =~ 1*x4 + x5 + x6 speed =~ 1*x7 + x8 + x9 # factor covariances visual ~~ textual + speed textual ~~ speed' fit <- lavaan(fit.mixed, data = HolzingerSwineford1939, auto.var = TRUE)
The best method to view results from a SEM fitted with lavaan is summary(), which can be called with optional arguments like fit.measures, standardized, and rsquare.
Core Lavaan Methods
summary() print a long summary of the model results
show() print a short summary of the model results
coef() returns the estimates of the free parameters in the model as a named numeric vector
fitted() returns the implied moments (covariance matrix and mean vector) of the model
resid() returns the raw, normalized or standardized residuals (difference between implied and observed moments)
vcov() returns the covariance matrix of the estimated parameters
predict() compute factor scores
logLik() returns the log-likelihood of the fitted model (if maximum likelihood estimation was used)
AIC(), BIC() compute information criteria (if maximum likelihood estimation is used)
update() update a fitted lavaan object
inspect() peek into the internal representation of the model; by default, it returns a list of model matrices counting the free parameters in the model; can also be used to extract starting values, gradient values, and much more
If these args are set to TRUE, the output includes additional fit measures, standardized estimates, and R2 values for the dependent variables, respectively
fit.model <- 'visual =~ x1 + x2 + x3 textual =~ x4 + x5 + x6 speed =~ x7 + x8 + x9'
fit <- cfa(fit.model, data = HolzingerSwineford1939)
summary(fit, fit.measures = TRUE)
fit <- cfa(fit.model, data=HolzingerSwineford1939, estimator="GLS", group="sex") fit.4 <- cfa(fit.model, data=HolzingerSwineford1939, estimator="GLD", group="sex", group.equal="regressions") anova(fit, fit.4)
The output consists of three sections.
- The first section (first 6 lines) contains the package version number, an indication whether the model has converged (and in how many iterations), and the effective number of observations used in the analysis.
- The second section contains the model χ^2 test statistic, degrees of freedom, and a p value are printed. If fit.measures = TRUE, it also prints the test statistic of the baseline model (where all observed variables are assumed to be uncorrelated) and several popular fit indices. If maximum likelihood estimation is used, this section will also contain information about the log-likelihood, the AIC, and the BIC.
- The third section provides an overview of the parameter estimates, including the type of standard errors used and whether the observed or expected information matrix was used to compute standard errors. Then, for each model parameter, the estimate and the standard error are displayed, and if appropriate, a z value based on the Wald test and a corresponding two-sided p value are also shown. To ease the reading of the parameter estimates, they are grouped into three blocks:
o factor loadings,
o factor covariances, and
o residual variances of both observed variables and factors.
The summary() method provides a nice summary of the model results for visualization purposes. The parameterEstimates() method returns the actual parameter estimates as a data.frame, which can be processed further.
| Index | lhs | op | rhs | est | se | z | pvalue | ci.lower | ci.upper |
| 1 | visual | =~ | x1 | 1 | 0 | NA | NA | 1 | 1 |
| 2 | visual | =~ | x2 | 0.553 | 0.1 | 5.554 | 0 | 0.358 | 0.749 |
| 3 | visual | =~ | x3 | 0.729 | 0.109 | 6.685 | 0 | 0.516 | 0.943 |
| 4 | textual | =~ | x4 | 1 | 0 | NA | NA | 1 | 1 |
| 5 | textual | =~ | x5 | 1.113 | 0.065 | 17.014 | 0 | 0.985 | 1.241 |
| 6 | textual | =~ | x6 | 0.926 | 0.055 | 16.703 | 0 | 0.817 | 1.035 |
| 7 | speed | =~ | x7 | 1 | 0 | NA | NA | 1 | 1 |
| 8 | speed | =~ | x8 | 1.18 | 0.165 | 7.152 | 0 | 0.857 | 1.503 |
| 9 | speed | =~ | x9 | 1.082 | 0.151 | 7.155 | 0 | 0.785 | 1.378 |
| 10 | x1 | ~~ | x1 | 0.549 | 0.114 | 4.833 | 0 | 0.326 | 0.772 |
| 11 | x2 | ~~ | x2 | 1.134 | 0.102 | 11.146 | 0 | 0.934 | 1.333 |
| 12 | x3 | ~~ | x3 | 0.844 | 0.091 | 9.317 | 0 | 0.667 | 1.022 |
| 13 | x4 | ~~ | x4 | 0.371 | 0.048 | 7.779 | 0 | 0.278 | 0.465 |
| 14 | x5 | ~~ | x5 | 0.446 | 0.058 | 7.642 | 0 | 0.332 | 0.561 |
| 15 | x6 | ~~ | x6 | 0.356 | 0.043 | 8.277 | 0 | 0.272 | 0.441 |
| 16 | x7 | ~~ | x7 | 0.799 | 0.081 | 9.823 | 0 | 0.64 | 0.959 |
| 17 | x8 | ~~ | x8 | 0.488 | 0.074 | 6.573 | 0 | 0.342 | 0.633 |
| 18 | x9 | ~~ | x9 | 0.566 | 0.071 | 8.003 | 0 | 0.427 | 0.705 |
| 19 | visual | ~~ | visual | 0.809 | 0.145 | 5.564 | 0 | 0.524 | 1.094 |
| 20 | textual | ~~ | textual | 0.979 | 0.112 | 8.737 | 0 | 0.76 | 1.199 |
| 21 | speed | ~~ | speed | 0.384 | 0.086 | 4.451 | 0 | 0.215 | 0.553 |
| 22 | visual | ~~ | textual | 0.408 | 0.074 | 5.552 | 0 | 0.264 | 0.552 |
| 23 | visual | ~~ | speed | 0.262 | 0.056 | 4.66 | 0 | 0.152 | 0.373 |
| 24 | textual | ~~ | speed | 0.173 | 0.049 | 3.518 | 0 | 0.077 | 0.27 |
The confidence level can be changed by setting the level argument. To obtain several standardized versions of the estimates, we can use standardized = TRUE:
subset(est, op == "=~")
| Index | lhs | op | rhs | est | se | z | pvalue | std.lv | std.all | std.nox |
| 1 | visual | =~ | x1 | 1 | 0 | NA | NA | 0.9 | 0.772 | 0.772 |
| 2 | visual | =~ | x2 | 0.553 | 0.1 | 5.554 | 0 | 0.498 | 0.424 | 0.424 |
| 3 | visual | =~ | x3 | 0.729 | 0.109 | 6.685 | 0 | 0.656 | 0.581 | 0.581 |
| 4 | textual | =~ | x4 | 1 | 0 | NA | NA | 0.99 | 0.852 | 0.852 |
| 5 | textual | =~ | x5 | 1.113 | 0.065 | 17.014 | 0 | 1.102 | 0.855 | 0.855 |
| 6 | textual | =~ | x6 | 0.926 | 0.055 | 16.703 | 0 | 0.917 | 0.838 | 0.838 |
| 7 | speed | =~ | x7 | 1 | 0 | NA | NA | 0.619 | 0.57 | 0.57 |
| 8 | speed | =~ | x8 | 1.18 | 0.165 | 7.152 | 0 | 0.731 | 0.723 | 0.723 |
| 9 | speed | =~ | x9 | 1.082 | 0.151 | 7.155 | 0 | 0.67 | 0.665 | 0.665 |
This only shows the factor loadings are shown but 3 additional columns with standardized values are added.
• In the first column (std.lv), only the latent variables have been standardized;
• In the second column (std.all), both the latent and the observed variables have been standardized;
• In the third column (std.nox), both the latent and the observed variables have been standardized, except for the exogenous observed variables. This option may be useful if the standardization of exogenous observed variables has little meaning (for example, binary covariates). Since there are no exogenous covariates in this model, the last two columns are identical in this output.
library("semPlot")
# semPaths(fit, "std", "show")
semPaths(fit, "std", curvePivot = TRUE, edge.label.cex = 1.0)
# get the margines right:
# semPaths(fit, "std", curvePivot = TRUE, edge.label.cex = 1.0, mar = c(10, 3, 10, 3))
# semPaths(fit, "std", curvePivot = TRUE, edge.label.cex = 1.0, mar = c(10, 3, 10, 3), as.expression = c("nodes",
# "edges"), sizeMan = 3, sizeInt = 1, sizeLat = 4)
Hands-on Example 2 (Parkinson’s Disease data)
# Data: PPMI Integrated (imaging, demographics, genetics, clinical and cognitive (UPDRS) data. # Dinov et al., 2015
INSERT CHART HERE
# install.packages("lavaan")
library(lavaan)
#load data 05_PPMI_top_UPDRS_Integrated_LongFormat1.csv ( dim(myData) 1764 31 )
# setwd("/dir/")
myData <- read.csv("https://umich.instructure.com/files/330397/download?download_frd=1&verifier=3bYRT9FXgBGMCQv8MNxsclWnMgodiJRYo3ODFtDq",header=TRUE)
# dichotomize the "ResearchGroup" variable myData$\$$ResearchGroup <- ifelse(myData$\$$ResearchGroup == "Control", 1, 0)
# Data elements: Index FID_IID L_cingulate_gyrus_ComputeArea L_cingulate_gyrus_Volume R_cingulate_gyrus_ComputeArea R_cingulate_gyrus_Volume L_caudate_ComputeArea L_caudate_Volume R_caudate_ComputeArea R_caudate_Volume L_putamen_ComputeArea L_putamen_Volume R_putamen_ComputeArea R_putamen_Volume L_hippocampus_ComputeArea L_hippocampus_Volume R_hippocampus_ComputeArea R_hippocampus_Volume cerebellum_ComputeArea cerebellum_Volume L_fusiform_gyrus_ComputeArea L_fusiform_gyrus_Volume R_fusiform_gyrus_ComputeArea R_fusiform_gyrus_Volume Sex Weight ResearchGroup Age chr12_rs34637584_GT chr17_rs11868035_GT chr17_rs11012_GT chr17_rs393152_GT chr17_rs12185268_GT UPDRS_Part_I_Summary_Score_Baseline UPDRS_Part_I_Summary_Score_Month_03 UPDRS_Part_I_Summary_Score_Month_06 UPDRS_Part_I_Summary_Score_Month_09 UPDRS_Part_I_Summary_Score_Month_12 UPDRS_Part_I_Summary_Score_Month_18 UPDRS_Part_I_Summary_Score_Month_24 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Baseline UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_09 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_12 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24 UPDRS_Part_III_Summary_Score_Baseline UPDRS_Part_III_Summary_Score_Month_03 UPDRS_Part_III_Summary_Score_Month_06 UPDRS_Part_III_Summary_Score_Month_09 UPDRS_Part_III_Summary_Score_Month_12 UPDRS_Part_III_Summary_Score_Month_18 UPDRS_Part_III_Summary_Score_Month_24 X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Baseline X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06 X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12 X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_24 X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06 X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_12 X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24
Validation of the measurement model
myData<-within(myData, {
L_cingulate_gyrus_ComputeArea <- lm(L_cingulate_gyrus_ComputeArea ~ L_cingulate_gyrus_Volume+R_cingulate_gyrus_ComputeArea+R_cingulate_gyrus_Volume+L_caudate_ComputeArea+L_caudate_Volume+R_caudate_ComputeArea+R_caudate_Volume+L_putamen_ComputeArea+L_putamen_Volume+R_putamen_ComputeArea+R_putamen_Volum e+L_hippocampus_ComputeArea+L_hippocampus_Volume+R_hippocampus_ComputeArea+R_hippocampus_Volume+cerebellum_ComputeArea+cerebellum_Volume+L_fusiform_gyrus_ComputeArea+L_fusiform_gyrus_Volume+R_fusiform_gyrus_ComputeArea+R_fusiform_gyru s_Volume, data=myData)$\$$residuals
Weight <- lm(Weight ~ Sex+ResearchGroup+Age+chr12_rs34637584_GT+chr17_rs11868035_GT+chr17_rs11012_GT+chr17_rs393152_GT+chr17_rs12185268_GT, data=myData)$\$$residuals
UPDRS_Part_I_Summary_Score_Baseline <- lm(UPDRS_Part_I_Summary_Score_Baseline ~ UPDRS_Part_I_Summary_Score_Month_03+UPDRS_Part_I_Summary_Score_Month_06+UPDRS_Part_I_Summary_Score_Month_09+UPDRS_Part_I_Summary_Score_Month_12+UPDRS_Part_I_Summary_Score_Month_18+UPDRS_Part_I_Summary_Score_Month_24+UPDRS_Part_II_Pati ent_Questionnaire_Summary_Score_Baseline+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_09+UPDRS_Part_II_Pa tient_Questionnaire_Summary_Score_Month_12+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24+UPDRS_Part_III_Summary_Score_Baseline+UPDRS_Part_III_Summary_Score_Month_ 03+UPDRS_Part_III_Summary_Score_Month_06+UPDRS_Part_III_Summary_Score_Month_09+UPDRS_Part_III_Summary_Score_Month_12+UPDRS_Part_III_Summary_Score_Month_18+UPDRS_Part_III_Summary_Score_Month_24+X_Assessment_Non.Motor_Epworth_Sleepiness _Scale_Summary_Score_Baseline+X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06+X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12+X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_ Month_24+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short _Summary_Score_Month_12+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24, data=myData)$\$$residuals
})
Structural Model
# Next, proceed with the structural model including the residuals from data to account for effects of site.
Lavaan model specification:
formula type operator mnemonic
latent variable definition =~ is measured by
regression ~ is regressed on
(residual) (co)variance ~~ is correlated with
Intercept ~ 1 Intercept
For example,
myModel <-
<b># regressions</b>
y1 + y2 <mark>~</mark> f1 + f2 + x1 + x2
f1 ~ f2 + f3
f2 ~ f3 + x1 + x2
<b># latent variable definitions</b>
f1 <mark>=~</mark> y1 + y2 + y3
f2 =~ y4 + y5 + y6
f3 =~ y7 + y8 + y9 + y10
<b># variances and covariances</b>
y1 <mark>~~</mark> y1
y1 ~~ y2
f1 ~~ f2
<b># intercepts</b>
y1 <mark>~</mark> 1
f1 ~ 1
model1 <-
'
# latent variable definitions - defining how the latent variables are “manifested by” a set of observed
# (or manifest) variables, aka “indicators”
# (1) Measurement Model
Imaging =~ L_cingulate_gyrus_ComputeArea+L_cingulate_gyrus_Volume
DemoGeno =~ Weight+Sex+Age
UPDRS =~ UPDRS_Part_I_Summary_Score_Baseline+UPDRS_Part_I_Summary_Score_Month_03
# (2) Regressions
ResearchGroup ~ Imaging + DemoGeno + UPDRS
'
model2 <-
'
# latent variable definitions - defining how the latent variables are “manifested by” a set of observed
# (or manifest) variables, aka “indicators”
# (1) Measurement Model
Imaging =~ L_cingulate_gyrus_ComputeArea+L_cingulate_gyrus_Volume+R_cingulate_gyrus_ComputeArea+R_cingulate_gyrus_Volume+L_caudate_ComputeArea+L_caudate_Volume+R_caudate_ComputeArea+R_caudate_Volume+L_putamen_ComputeArea+L_putamen_Volume+R_putam en_ComputeArea+R_putamen_Volume+L_hippocampus_ComputeArea+L_hippocampus_Volume+R_hippocampus_ComputeArea+R_hippocampus_Volume+cerebellum_ComputeArea+cerebellum_Volume+L_fusiform_gyrus_ComputeArea+L_fusiform_gyrus_Volume+R_fusiform_gyr us_ComputeArea+R_fusiform_gyrus_Volume
DemoGeno =~ Weight+Sex+Age+chr12_rs34637584_GT+chr17_rs11868035_GT+chr17_rs11012_GT+chr17_rs393152_GT+chr17_rs12185268_GT
UPDRS =~ UPDRS_Part_I_Summary_Score_Baseline+UPDRS_Part_I_Summary_Score_Month_03+UPDRS_Part_I_Summary_Score_Month_06+UPDRS_Part_I_Summary_Score_Month_09+UPDRS_Part_I_Summary_Score_Month_12+UPDRS_Part_I_Summary_Score_Month_18+UPDRS_Part_I_Summa ry_Score_Month_24+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Baseline+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06+UPDRS_Part_II_Patient_Questionnaire_Sum mary_Score_Month_09+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_12+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18+UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24+UPDRS_Part_III_Summary_Score_Baseline +UPDRS_Part_III_Summary_Score_Month_03+UPDRS_Part_III_Summary_Score_Month_06+UPDRS_Part_III_Summary_Score_Month_09+UPDRS_Part_III_Summary_Score_Month_12+UPDRS_Part_III_Summary_Score_Month_18+UPDRS_Part_III_Summary_Score_Month_24+X_Ass essment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Baseline+X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06+X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12+X_Assessment_Non.Motor_Epw orth_Sleepiness_Scale_Summary_Score_Month_24+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06+X_Assessment_Non.Motor_ Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_12+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24
# (2) Regressions
# ResearchGroup ~ Imaging + DemoGeno + UPDRS
# transform cat variable to numeric:
# myData$\$$ResearchGroup <- ifelse(myData$\$$ResearchGroup == "Control", 0,
# ifelse(myData$\$$ResearchGroup == "PD", 2, 1))
RG_ranked ~ Imaging + DemoGeno + UPDRS
# (3) Residual Variances L_insular_cortex_ComputeArea ~~ L_insular_cortex_ComputeArea L_insular_cortex_Volume ~~ L_insular_cortex_Volume R_insular_cortex_ComputeArea ~~ R_insular_cortex_ComputeArea R_insular_cortex_Volume ~~ R_insular_cortex_Volume L_cingulate_gyrus_ComputeArea ~~ L_cingulate_gyrus_ComputeArea L_cingulate_gyrus_Volume ~~ L_cingulate_gyrus_Volume R_cingulate_gyrus_ComputeArea ~~ R_cingulate_gyrus_ComputeArea R_cingulate_gyrus_Volume ~~ R_cingulate_gyrus_Volume L_caudate_ComputeArea ~~ L_caudate_ComputeArea L_caudate_Volume ~~ L_caudate_Volume R_caudate_ComputeArea ~~ R_caudate_ComputeArea R_caudate_Volume ~~ R_caudate_Volume L_putamen_ComputeArea ~~ L_putamen_ComputeArea L_putamen_Volume ~~ L_putamen_Volume R_putamen_ComputeArea ~~ R_putamen_ComputeArea R_putamen_Volume ~~ R_putamen_Volume L_hippocampus_ComputeArea ~~ L_hippocampus_ComputeArea L_hippocampus_Volume ~~ L_hippocampus_Volume R_hippocampus_ComputeArea ~~ R_hippocampus_ComputeArea R_hippocampus_Volume ~~ R_hippocampus_Volume cerebellum_ComputeArea ~~ cerebellum_ComputeArea cerebellum_Volume ~~ cerebellum_Volume L_fusiform_gyrus_ComputeArea ~~ L_fusiform_gyrus_ComputeArea L_fusiform_gyrus_Volume ~~ L_fusiform_gyrus_Volume R_fusiform_gyrus_ComputeArea ~~ R_fusiform_gyrus_ComputeArea R_fusiform_gyrus_Volume ~~ R_fusiform_gyrus_Volume R_fusiform_gyrus_ShapeIndex ~~ R_fusiform_gyrus_ShapeIndex R_fusiform_gyrus_Curvedness ~~ R_fusiform_gyrus_Curvedness Sex ~~ Sex Weight ~~ Weight ResearchGroup ~~ ResearchGroup VisitID ~~ VisitID Age ~~ Age chr12_rs34637584_GT ~~ chr12_rs34637584_GT chr17_rs11868035_GT ~~ chr17_rs11868035_GT chr17_rs11012_GT ~~ chr17_rs11012_GT chr17_rs393152_GT ~~ chr17_rs393152_GT chr17_rs12185268_GT ~~ chr17_rs12185268_GT chr17_rs199533_GT ~~ chr17_rs199533_GT UPDRS_Part_I_Summary_Score_Baseline ~~ UPDRS_Part_I_Summary_Score_Baseline UPDRS_Part_I_Summary_Score_Month_03 ~~ UPDRS_Part_I_Summary_Score_Month_03 UPDRS_Part_I_Summary_Score_Month_06 ~~ UPDRS_Part_I_Summary_Score_Month_06 UPDRS_Part_I_Summary_Score_Month_09 ~~ UPDRS_Part_I_Summary_Score_Month_09 UPDRS_Part_I_Summary_Score_Month_12 ~~ UPDRS_Part_I_Summary_Score_Month_12 UPDRS_Part_I_Summary_Score_Month_18 ~~ UPDRS_Part_I_Summary_Score_Month_18 UPDRS_Part_I_Summary_Score_Month_24 ~~ UPDRS_Part_I_Summary_Score_Month_24 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Baseline ~~ UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Baseline UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03 ~~ UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06 ~~ UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_09 ~~ UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_09 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_12 ~~ UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_12 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18 ~~ UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18 UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24 ~~ UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24 UPDRS_Part_III_Summary_Score_Baseline ~~ UPDRS_Part_III_Summary_Score_Baseline UPDRS_Part_III_Summary_Score_Month_03 ~~ UPDRS_Part_III_Summary_Score_Month_03 UPDRS_Part_III_Summary_Score_Month_06 ~~ UPDRS_Part_III_Summary_Score_Month_06 UPDRS_Part_III_Summary_Score_Month_09 ~~ UPDRS_Part_III_Summary_Score_Month_09 UPDRS_Part_III_Summary_Score_Month_12 ~~ UPDRS_Part_III_Summary_Score_Month_12 UPDRS_Part_III_Summary_Score_Month_18 ~~ UPDRS_Part_III_Summary_Score_Month_18 UPDRS_Part_III_Summary_Score_Month_24 ~~ UPDRS_Part_III_Summary_Score_Month_24 X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Baseline ~~ X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Baseline X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06 ~~ X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06 X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12 ~~ X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12 X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_24 ~~ X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_24 X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline ~~ X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06 ~~ X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06 X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_12 ~~ X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_12 X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24 ~~ X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24
# (4) Residual Covariances Sex ~~ Weight '
# confirmatory factor analysis (CFA) # The baseline is a null model constraining the observed variables to covary with no other variables. # That is, the covariances are fixed to 0 and only individual variances are estimated. This is represents # a “reasonable worst-possible fitting model”, against which the new fitted model is compared # to calculate appropriate model-quality indices (e.g., CFA).
# standardize all variable to avoid huge variations between variable distributions
library("MASS")
# myData <- read.csv("https://umich.instructure.com/files/330397/download?download_frd=1&verifier=3bYRT9FXgBGMCQv8MNxsclWnMgodiJRYo3ODFtDq",header=TRUE)
summary(myData) myData2<-scale(myData); summary(myData2)
myDF <- data.frame(myData2)
# myDF3 <- subset(myDF, select=c("L_cingulate_gyrus_ComputeArea", "cerebellum_Volume", "Weight", "Sex", "Age", " UPDRS_part_I", "UPDRS_part_II", "UPDRS_part_III", "ResearchGroup"))
myDF3 <- subset(myDF, select=c("R_insular_cortex_ComputeArea", "R_insular_cortex_Volume", "Sex", "Weight", "ResearchGroup", "Age", "chr12_rs34637584_GT", "chr17_rs11868035_GT", "chr17_rs11012_GT"))
model3 <- ' # latent variable definitions - defining how the latent variables are “manifested by” a set of observed # (or manifest) variables, aka “indicators” # (1) Measurement Model # Imaging =~ L_cingulate_gyrus_ComputeArea + cerebellum_Volume Imaging =~ R_insular_cortex_ComputeArea + R_insular_cortex_Volume DemoGeno =~ Weight+Sex+Age # UPDRS =~ UPDRS_Part_I_Summary_Score_Baseline+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline UPDRS =~ UPDRS_part_I +UPDRS_part_II + UPDRS_part_III # (2) Regressions ResearchGroup ~ Imaging + DemoGeno + UPDRS
fit3 <- cfa(model3, data= myData2, missing='FIML') # deal with missing values (missing='FIML') summary(fit3, fit.measures=TRUE)
lavaan (0.5-18) converged normally after 2044 iterations
Number of observations 1764
Number of missing patterns 3
Estimator ML
Minimum Function Test Statistic 455.923
Degrees of freedom 15
P-value (Chi-square) 0.000
Model test baseline model:
Minimum Function Test Statistic 2625.020
Degrees of freedom 28
P-value 0.000
User model versus baseline model:
Comparative Fit Index (CFI) 0.830
Tucker-Lewis Index (TLI) 0.683
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -51499.484
Loglikelihood unrestricted model (H1) -51271.522
Number of free parameters 29
Akaike (AIC) 103056.967
Bayesian (BIC) 103215.752
Sample-size adjusted Bayesian (BIC) 103123.621
Root Mean Square Error of Approximation:
RMSEA 0.129
90 Percent Confidence Interval 0.119 0.139
P-value RMSEA <= 0.05 0.000
Standardized Root Mean Square Residual:
SRMR 0.062
Parameter estimates:
Information Observed
Standard Errors Standard
Estimate Std.err Z-value P(>|z|)
Latent variables:
Imaging =~
R_cnglt_gyr_V 1.000
L_cadt_CmptAr 493.058
DemoGeno =~
Weight 1.000
Sex 24.158
Age 0.094
UPDRS =~
UPDRS_part_I 1.000
UPDRS_part_II 7.389
Regressions:
ResearchGroup ~
Imaging -0.000
DemoGeno 0.002
UPDRS -0.323
Covariances:
Imaging ~~
DemoGeno 0.001
UPDRS 0.002
DemoGeno ~~
UPDRS 0.000
Intercepts:
R_cnglt_gyr_V 7895.658
L_cadt_CmptAr 635.570
Weight 82.048
Sex 1.340
Age 61.073
UPDRS_part_I 1.126
UPDRS_part_II 4.905
ResearchGroup 0.290
Imaging 0.000
DemoGeno 0.000
UPDRS 0.000
Variances:
R_cnglt_gyr_V 17070159.189
L_cadt_CmptAr -536243845.090
Weight 274.912
Sex 96.664
Age 105.347
UPDRS_part_I 2.442
UPDRS_part_II -0.256
ResearchGroup 0.149
Imaging 2206.397
DemoGeno -0.165
UPDRS 0.550
3 parts of the Lavaan SEM output
• First six lines are called the header contains the following information:
• lavaan version number
• lavaan converge info (normal or not), and # iterations needed
• the number of observations that were effectively used in the analysis
• the estimator that was used to obtain the parameter values (here: ML)
• the model test statistic, the degrees of freedom, and a corresponding p-value
(1) Next, is the Model test baseline model and the value for the SRMR
(2) The last section contains the parameter estimates, standard errors (if the information matrix is expected or observed, and if the standard errors are standard, robust, or based on the bootstrap). Then, it tabulates all free (and fixed) parameters that were included in the model. Typically, first the latent variables are shown, followed by covariances and (residual) variances. The first column (Estimate) contains the (estimated or fixed) parameter value for each model parameter; the second column (Std.err) contains the standard error for each estimated parameter; the third column (Z-value) contains the Wald statistic (which is simply obtained by dividing the parameter value by its standard error), and the last column contains the p-value for testing the null hypothesis that the parameter equals zero in the population.
Note: You can get this type of error “…system is computationally singular: reciprocal condition…”, which indicates that the design matrix is not invertible. Thus, it can't be used to develop a regression model. This is due to linearly dependent columns, i.e. strongly correlated variables. Resolve pairwise covariances (or correlations) of your variables to investigate if there are any variables that can potentially be removed. You're looking for covariances (or correlations) >> 0. We can also automate this variable selection by using a forward stepwise regression.
# Graphical fit model visualization library(semPlot) semPaths(fit3)
semPaths(fit3, "std", ask = FALSE, as.expression = "edges", mar = c(3, 1, 5, 1))
sem() vs. cfa()
The function sem() is very similar to the function cfa(). As we did not include fit.measures=TRUE, the report only includes the basic chi-square test statistic. The argument standardized=TRUE, reports standardized parameter values. Two extra columns of standardized parameter values are printed. In the first column (labeled, Std.lv), only the latent variables are standardized. In the second column (labeled Std.all), both latent and observed variables are standardized. The latter is often called the `completely standardized solution'.
# remove all objects from the R console (current workspace) rm(list = ls())
# library("lavaan")
#load data 05_PPMI_top_UPDRS_Integrated_LongFormat1.csv ( dim(myData) 1764 31 ), long format
# myData <- read.csv("https://umich.instructure.com/files/330397/download?download_frd=1&verifier=3bYRT9FXgBGMCQv8MNxsclWnMgodiJRYo3ODFtDq",header=TRUE)
# dichotomize the "ResearchGroup" variable
myData$\$$ResearchGroup <- ifelse(myData$\$$ResearchGroup == "Control", 1, 0)
# library("MASS")
myData2<-scale(myData); head(myData2)
myData2[, 20] <- myData$\$$ResearchGroup; # replace the ResearchGroup by orig binary labels (do not rescale)
attach(myData2)
head(myData2)
# model3 <-
'
# latent variable definitions - defining how the latent variables are “manifested by” a set of observed
# (or manifest) variables, aka “indicators”
# (1) Measurement Model
# Imaging =~ L_cingulate_gyrus_ComputeArea + cerebellum_Volume
Imaging =~ R_insular_cortex_ComputeArea + R_insular_cortex_Volume
DemoGeno =~ Weight+Sex+Age
# UPDRS =~ UPDRS_Part_I_Summary_Score_Baseline+X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline
UPDRS =~ UPDRS_part_I +UPDRS_part_II + UPDRS_part_III
# (2) Regressions
ResearchGroup ~ Imaging + DemoGeno + UPDRS
'
Here is why we needed to scale the data first before we fit the SEM model:
# using raw data provides unreliable estimates
<font color="red"># some observed variances are (at least) a factor 1000 times larger than others;</font>
fit3 <- sem(model3, data=<b>myData</b>, estimator="MLM")
summary(fit3)
# using scaled data provides stable estimates
fit3 <- sem(model3, data=myData2, estimator="MLM")
summary(fit3)
# report the standardized coefficients of the model
standardizedSolution(fit3)
# variation explained by model components (The R-square value for all endogenous variables)
inspect(fit3, "r2")
Note that all variances are supposed to be positive, however, occasionally, model estimates may generate a residual variance that is negative. This may happen due to random sampling error where a very small true value may sometimes produce a negative estimate, or it can occur when the model is unstable.
# Inspect the fitted values variance-covariance matrix:
fitted(fit3)$\$$cov
#
Model fitting
Name Command
fit CFA to data cfa(model, data=Data)
fit SEM to data sem(model, data=Data)
standardized solution sem(model, data=Data, std.ov=TRUE)
orthogonal factors cfa(model, data=Data, orthogonal=TRUE)
Matrices
Name Command
Factor covariance matrix inspect(fit, "coefficients")$\$$psi
Fitted covariance matrix fitted(fit)$\$$cov
Observed covariance matrix inspect(fit, 'sampstat')$\$$cov
Residual covariance matrix resid(fit)$\$$cov
Factor correlation matrix cov2cor(inspect(fit, "coefficients")$\$$psi) or use covariance command with standardised solution e.g., cfa(..., std.ov=TRUE)
Fit Measures
<b>Name Command</b>
Fit measures: fitMeasures(fit)
Specific fit measures e.g.: fitMeasures(fit)[c('chisq', 'df', 'pvalue', 'cfi', 'rmsea', 'srmr')]
Parameters
<b>Name Command</b>
Parameter information parTable(fit)
Standardised estimates standardizedSolution(fit) or summary(fit, standardized=TRUE)
R-squared | inspect(fit, 'r2')
Compare models
<b>Name Command</b>
Compare fit measures cbind(m1=inspect(m1_fit, 'fit.measures'), m2=inspect(m2_fit, 'fit.measures'))
Chi-square difference test anova(m1_fit, m2_fit)
Model improvement
<b>Name Command</b>
Modification indices mod_ind <- modificationindices(fit)
10 greatest head(mod_ind[order(mod_ind$\$$mi, decreasing=TRUE), ], 10)
mi > 5 subset(mod_ind[order(mod_ind$\$$mi, decreasing=TRUE), ], mi > 5)
# to account for groupings (gender)
# fit3 <- sem(model3, data=myData, group="Sex", estimator="MLM")
# install.packages("semPlot")
library(semPlot)
# Plot standardized model (numerical):
# semPaths(fit3, what = "est", layout = "tree", title = TRUE, style = "LISREL")
semPaths(fit3)
[[Image:SMHS_BigDataBigSci7.png|500px]]
=='"`UNIQ--h-1--QINU`"'Growth Curve Models==
Latent growth curve models may be used to analyze longitudinal or temporal data where the outcome measure is assessed on multiple occasions, and we examine its change over time, e.g., the trajectory over time can be
modeled as a linear or quadratic function. Random effects are used to capture individual differences by conveniently representing (continuous) latent variables, aka growth factors. To fit a linear growth model we may specify a model with two latent variables: a random intercept, and a random slope:
#load data <b>05_PPMI_top_UPDRS_Integrated_LongFormat.csv ( dim(myData) 661 71), wide</b>
# setwd("/dir/")
myData <- read.csv("https://umich.instructure.com/files/330395/download?download_frd=1&verifier=v6jBvV4x94ka3EYcGKuXXg5BZNaOLBVp0xkJih0H",header=TRUE)
attach(myData)
# dichotomize the "ResearchGroup" variable
table(myData$\$$ResearchGroup)
myData$\$$ResearchGroup <- ifelse(myData$\$$ResearchGroup == "Control", 1, 0)
# linear growth model with 4 timepoints # intercept (i) and slope (s) with fixed coefficients # i =~ 1*t1 + 1*t2 + 1*t3 + 1*t4 (intercept/constant) # s =~ 0*t1 + 1*t2 + 2*t3 + 3*t4 (slope/linear term) # ??? =~ 0*t1 + 1*t2 + 2*t3 + 3*t4 (quadratic term)
In this model, we have fixed all the coefficients of the linear growth functions: model4 <-
'
i =~ 1*UPDRS_Part_I_Summary_Score_Baseline + 1*UPDRS_Part_I_Summary_Score_Month_03 + 1*UPDRS_Part_I_Summary_Score_Month_06 + 1*UPDRS_Part_I_Summary_Score_Month_09 + 1*UPDRS_Part_I_Summary_Score_Month_12 + 1*UPDRS_Part_I_Summary_Score_Month_18 + 1*UPDRS_Part_I_Summary_Score_Month_24 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Baseline + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_09 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_12 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18 + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24 + 1*UPDRS_Part_III_Summary_Score_Baseline + 1*UPDRS_Part_III_Summary_Score_Month_03 + 1*UPDRS_Part_III_Summary_Score_Month_06 + 1*UPDRS_Part_III_Summary_Score_Month_09 + 1*UPDRS_Part_III_Summary_Score_Month_12 + 1*UPDRS_Part_III_Summary_Score_Month_18 + 1*UPDRS_Part_III_Summary_Score_Month_24 + 1*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Baseline + 1*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06 + 1*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12 + 1*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_24 + 1*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline + 1*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06 + 1*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_12 + 1*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24 s =~ 0*UPDRS_Part_I_Summary_Score_Baseline + 1*UPDRS_Part_I_Summary_Score_Month_03 + 2*UPDRS_Part_I_Summary_Score_Month_06 + 3*UPDRS_Part_I_Summary_Score_Month_09 + 4*UPDRS_Part_I_Summary_Score_Month_12 + 5*UPDRS_Part_I_Summary_Score_Month_18 + 6*UPDRS_Part_I_Summary_Score_Month_24 + 0*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Baseline + 1*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_03 + 2*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_06 + 3*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_09 + 4*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_12 + 5*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_18 + 6*UPDRS_Part_II_Patient_Questionnaire_Summary_Score_Month_24 + 0*UPDRS_Part_III_Summary_Score_Baseline + 1*UPDRS_Part_III_Summary_Score_Month_03 + 2*UPDRS_Part_III_Summary_Score_Month_06 + 3*UPDRS_Part_III_Summary_Score_Month_09 + 4*UPDRS_Part_III_Summary_Score_Month_12 + 5*UPDRS_Part_III_Summary_Score_Month_18 + 6*UPDRS_Part_III_Summary_Score_Month_24 + 0*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Baseline + 2*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_06 + 4*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_12 + 6*X_Assessment_Non.Motor_Epworth_Sleepiness_Scale_Summary_Score_Month_24 + 0*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Baseline + 2*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_06 + 4*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_12 + 6*X_Assessment_Non.Motor_Geriatric_Depression_Scale_GDS_Short_Summary_Score_Month_24
fit4 <- growth(model4, data=myData) summary(fit4) parameterEstimates(fit4) # extracts the values of the estimated parameters, the standard errors, # the z-values, the standardized parameter values, and returns a data frame fitted(fit4) # return the model-implied (fitted) covariance matrix (and mean vector) of a fitted model
# resid() function return (unstandardized) residuals of a fitted model including the difference between # the observed and implied covariance matrix and mean vector resid(fit4)
Measures of model quality (Comparative Fit Index (CFI), Root Mean Square Error of Approximation (RMSEA))
# report the fit measures as a signature vector: Comparative Fit Index (CFI), Root Mean Square Error of
# Approximation (RMSEA)
fitMeasures(fit4, c("cfi", "rmsea", "srmr"))
Comparative Fit Index (CFI) is an incremental measure directly based on the non-centrality measure. If d = χ2(df) where df are the degrees of freedom of the model, the Comparative Fit Index is:
FIX THIS!!!!!!!!!!!!!!!!(d(Null Model) - d(Proposed Model))/(d(Null Model)).
0≤CFI≤1 (by definition). It is interpreted as:
- CFI<0.9 - model fitting is poor.
- 0.9≤CFI≤0.95 is considered marginal,
- CFI>0.95 is good.
CFI is a relative index of model fit – it compare the fit of your model to the fit of (the worst) fitting null model.
Root Mean Square Error of Approximation (RMSEA) - “Ramsey”
An absolute measure of fit based on the non-centrality parameter: FIX EQUATION!!!!>√((χ2 - df)/(df×(N - 1))) ,
where N the sample size and df the degrees of freedom of the model. If χ2 < df, then the RMSEA∶=0. It has a penalty for complexity via the chi square to df ratio. The RMSEA is a popular measure of model fit.
- RMSEA < 0.01, excellent,
- RMSEA < 0.05, good
- RMSEA > 0.10 cutoff for poor fitting models
Standardized Root Mean Square Residual (SRMR) is an absolute measure of fit defined as the standardized difference between the observed correlation and the predicted correlation. A value of zero indicates perfect fit. The SRMR has no penalty for model complexity. SRMR <0.08 is considered a good fit.
# inspect the model results (report parameter table) inspect(fit4)
#install.packages("semTools")
# library("semTools")
A Simpler Model (fit5)
model5 <- '
# intercept and slope with fixed coefficients
i =~ UPDRS_Part_I_Summary_Score_Baseline + UPDRS_Part_I_Summary_Score_Month_03 + UPDRS_Part_I_Summary_Score_Month_24
s =~ 0*UPDRS_Part_I_Summary_Score_Baseline + 1*UPDRS_Part_I_Summary_Score_Month_03 + 6*UPDRS_Part_I_Summary_Score_Month_24
# regressions
i ~ R_fusiform_gyrus_Volume + Weight + ResearchGroup + Age + chr12_rs34637584_GT
s ~ R_fusiform_gyrus_Volume + Weight + ResearchGroup + Age + chr12_rs34637584_GT
# time-varying covariates
UPDRS_Part_I_Summary_Score_Baseline ~ Weight
UPDRS_Part_I_Summary_Score_Month_03 ~ ResearchGroup
UPDRS_Part_I_Summary_Score_Month_24 ~ Age
'
fit5 <- growth(model5, data=myData)
summary(fit5); fitMeasures(fit5, c("cfi", "rmsea", "srmr"))
parameterEstimates(fit5) # extracts the values of the estimated parameters, the standard errors,
# the z-values, the standardized parameter values, and returns a data frame
See also
- SOCR Home page: http://www.socr.umich.edu
Translate this page: