Difference between revisions of "SOCR Activity ANOVA FlignerKilleen MeatConsumption"
| Line 174: | Line 174: | ||
Open the [http://www.socr.ucla.edu/htmls/ana/ANOVA2Way_Analysis.html SOCR ANOVA-Two Way applet] (requires Java-enabled browser). | Open the [http://www.socr.ucla.edu/htmls/ana/ANOVA2Way_Analysis.html SOCR ANOVA-Two Way applet] (requires Java-enabled browser). | ||
| − | + | For the following analyses, we will focus on the data for Beef consumption. Let us test to see if different countries eat different amounts of beef. | |
| + | Usually when we compare a set of groups as this one, we would use a one-way ANOVA (comparing the seven countries). To run this test, open up the [[AP_Statistics_Curriculum_2007_ANOVA_1Way|one-way ANOVA analysis]] in the [http://www.socr.ucla.edu/htmls/ana/ANOVA1Way_Analysis.html SOCR Analyses Applet] in a java-enabled browser. It should be the default setting when you open up the page: | ||
| − | + | <center>[[Image:SOCR_Activity_ANOVA_FlignerKilleen_MeatConsumption_Fig6.png|500px]]</center> | |
| − | + | Now, prepare your dataset (it will be the Beef table from the above summary tables). We will be treating the yearly results as being our sample’s data points, attempting to capture the overall population consumption. This seems reasonable; the average meet consumption of a country should not change that much in a seven-year timespan. | |
| − | + | Once you have rearranged your dataset for use in the ANOVA applet (if you do not know what it should look like, try considering one of the SOCR ANOVA tutorials). It should look like this in the applet data screen: | |
| − | <center>[[Image: | + | <center>[[Image:SOCR_Activity_ANOVA_FlignerKilleen_MeatConsumption_Fig7.png|500px]]</center> |
| − | + | Rename your column headers to define the independent and dependent variables. | |
| − | : | + | <center>[[Image:SOCR_Activity_ANOVA_FlignerKilleen_MeatConsumption_Fig8.png|500px]]</center> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Click on the mapping tab, and assign your independent and dependent variables appropriately: | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | : | + | <center>[[Image:SOCR_Activity_ANOVA_FlignerKilleen_MeatConsumption_Fig9.png|500px]]</center> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | : | + | Set your precision to all, then click calculate: |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | : | + | <center>[[Image:SOCR_Activity_ANOVA_FlignerKilleen_MeatConsumption_Fig10.png|500px]]</center> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | : | + | The following results should appear in the '''Results''' tab: |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | : Sample Size = 49 | |
| + | : Independent Variable = Country | ||
| + | : Dependent Variable = Consumption | ||
| + | : Results of One-Way Analysis of Variance: | ||
| + | : [[AP_Statistics_Curriculum_2007_ANOVA_1Way|Standard 1-Way ANOVA Table]] | ||
| + | ============================================================================================== | ||
| + | VarianceSource DF RSS MSS F-Statistics P-value | ||
| + | TreatmentEffect (B/w Groups) 6 668292765.4285718000 111382127.5714286400 898.8918351858 < 1E-15 | ||
| + | Error 42 5204240.5714285690 123910.4897959183 | ||
| + | Total: 48 673497006.0000004000 | ||
| + | ============================================================================================== | ||
| + | |||
| + | : Model: | ||
| + | : Degrees of Freedom = 6 | ||
| + | : Residual Sum of Squares = 668292765.4285718000 | ||
| + | : Mean Square Error = 111382127.5714286400 | ||
| + | |||
| + | : Error: | ||
| + | : Degrees of Freedom = 42 | ||
| + | : Residual Sum of Squares = 5204240.5714285690 | ||
| + | : Mean Square Error = 123910.4897959183 | ||
| + | |||
| + | : Corrected Total: | ||
| + | : Degrees of Freedom = 48 | ||
| + | : Residual Sum of Squares = 673497006.0000004000 | ||
| + | |||
| + | : F-Value = 898.8918351858 | ||
| + | : P-Value = < 1E-15 | ||
| + | : R-Square = 0.9922728082 | ||
==Conclusions== | ==Conclusions== | ||
Revision as of 17:56, 21 February 2013
Contents
- 1 SOCR Educational Materials - Activities - SOCR Meat Consumption Activity – ANOVA assumptions about the variance homogeneity Activity
- 2 Motivation and Goals
- 3 Summary
- 4 Data
- 5 Exploratory data analyses (EDA)
- 6 Quantitative data analysis (QDA)
- 7 Conclusions
- 8 Practice problems
- 9 See also
- 10 References
SOCR Educational Materials - Activities - SOCR Meat Consumption Activity – ANOVA assumptions about the variance homogeneity Activity
Motivation and Goals
In many developed countries, when people imagine their next meal, they focus on one specific part: the meat. That choice of meat, however, varies from country to country due to the popularity and availability of various domesticated animals. Furthermore, the amount of meat eaten has a surprising degree of variability across time, cultures and geographic regions.
The following activity will study the effects of that variance on the statistical analyses. Specifically, we will consider how deviations from homoscedasticity (also known as equivalence of variance or variance homogeneity) can lead to making some incomplete or even incorrect conclusions. To do so, we will employ the Fligner-Killeen method to analyze some real meet consumption data.
Summary
This activity uses a reduced version of the open-source meat-consumption dataset. All data comes from the US Census Bureau.
This dataset summarizes the meat consumption, by animal type, of various countries (the European Union (EU) is being treated as a single country in this case). For simplicity, records from countries that did provide consumption measures for all meat types and all years were removed from the data set.
Data
Data Description
- Number of cases: 147
- Variables
- Country: The country or world region in question
- Brazil
- China
- European Union
- Japan
- Mexico
- Russia
- United States
- Meat: The type of meat
- Beef
- Pork
- Poultry
- Years Represented (2000 – 2006)
- Country: The country or world region in question
- Values are in thousands of metric tons
Data Summaries
Chicken/Poultry
| Year | Brazil | China | Europe | Japan | Mexico | Russia | UnitedStates | YearAverage | YearSD |
|---|---|---|---|---|---|---|---|---|---|
| 2000 | 5110 | 9393 | 6934 | 1772 | 2163 | 1320 | 11474 | 5452.286 | 3990.459 |
| 2001 | 5341 | 9237 | 7359 | 1797 | 2311 | 1588 | 11558 | 5598.714 | 3942.57 |
| 2002 | 5873 | 9556 | 7417 | 1830 | 2424 | 1697 | 12270 | 5866.714 | 4134.211 |
| 2003 | 5742 | 9963 | 7312 | 1841 | 2627 | 1680 | 12540 | 5957.857 | 4234.565 |
| 2004 | 5992 | 9931 | 7280 | 1713 | 2713 | 1675 | 13080 | 6054.857 | 4379.591 |
| 2005 | 6612 | 10088 | 7596 | 1880 | 2871 | 2139 | 13430 | 6373.714 | 4388.111 |
| 2006 | 6853 | 10371 | 7380 | 1908 | 3005 | 2382 | 13754 | 6521.857 | 4448.974 |
| Country_Average | 5931.857 | 9791.286 | 7325.429 | 1820.143 | 2587.714 | 1783 | 12586.57 | ||
| Country_SD | 629.6543 | 407.0908 | 200.4826 | 66.03895 | 304.2404 | 357.6777 | 886.5564 |
Pork
| Year | Brazil | China | Europe | Japan | Mexico | Russia | UnitedStates | YearAverage | YearSD |
|---|---|---|---|---|---|---|---|---|---|
| 2000 | 1827 | 40378 | 19242 | 2228 | 1252 | 2019 | 8455 | 10771.57 | 14570.99 |
| 2001 | 1919 | 41829 | 19317 | 2268 | 1298 | 2076 | 8389 | 11013.71 | 15049.33 |
| 2002 | 1975 | 43238 | 19746 | 2377 | 1349 | 2453 | 8685 | 11403.29 | 15502.7 |
| 2003 | 1957 | 45054 | 20043 | 2373 | 1423 | 2420 | 8816 | 11726.57 | 16145.49 |
| 2004 | 1979 | 46648 | 19773 | 2562 | 1556 | 2337 | 8817 | 11953.14 | 16648.16 |
| 2005 | 1949 | 49703 | 19768 | 2507 | 1556 | 2476 | 8669 | 12375.43 | 17714.83 |
| 2006 | 2191 | 51809 | 20015 | 2450 | 1580 | 2637 | 8640 | 12760.29 | 18438.64 |
| Country_Average | 1971 | 45522.71 | 19700.57 | 2395 | 1430.571 | 2345.429 | 8638.714 | ||
| Country_SD | 110 | 4159.521 | 312.355 | 121.3013 | 135.3808 | 223.0148 | 164.5121 |
Beef
| Year | Brazil | China | Europe | Japan | Mexico | Russia | UnitedStates | YearAverage | YearSD |
|---|---|---|---|---|---|---|---|---|---|
| 2000 | 6102 | 5284 | 8106 | 1585 | 2309 | 2246 | 12502 | 5447.714 | 3922.316 |
| 2001 | 6191 | 5434 | 7658 | 1419 | 2341 | 2400 | 12351 | 5399.143 | 3835.093 |
| 2002 | 6437 | 5818 | 8187 | 1319 | 2409 | 2450 | 12737 | 5622.429 | 4016.753 |
| 2003 | 6273 | 6274 | 8315 | 1366 | 2308 | 2378 | 12340 | 5607.714 | 3933.847 |
| 2004 | 6400 | 6703 | 8292 | 1182 | 2368 | 2308 | 12667 | 5702.857 | 4077.861 |
| 2005 | 6774 | 7026 | 8194 | 1200 | 2419 | 2503 | 12663 | 5825.571 | 4056.693 |
| 2006 | 6939 | 7395 | 8270 | 1173 | 2509 | 2370 | 12830 | 5926.571 | 4148.408 |
| Country_Average | 6445.143 | 6276.286 | 8146 | 1320.571 | 2380.429 | 2379.286 | 12584.29 | ||
| Country_SD | 307.1685 | 806.2036 | 226.9295 | 151.2138 | 71.75388 | 85.31259 | 190.4396 |
Raw Dataset
| Country | Meat | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 |
|---|---|---|---|---|---|---|---|---|
| Brazil | Beef | 6102 | 6191 | 6437 | 6273 | 6400 | 6774 | 6939 |
| Brazil | Pork | 1827 | 1919 | 1975 | 1957 | 1979 | 1949 | 2191 |
| Brazil | Poultry | 5110 | 5341 | 5873 | 5742 | 5992 | 6612 | 6853 |
| China | Beef | 5284 | 5434 | 5818 | 6274 | 6703 | 7026 | 7395 |
| China | Pork | 40378 | 41829 | 43238 | 45054 | 46648 | 49703 | 51809 |
| China | Poultry | 9393 | 9237 | 9556 | 9963 | 9931 | 10088 | 10371 |
| EuropeanUnion | Beef | 8106 | 7658 | 8187 | 8315 | 8292 | 8194 | 8270 |
| EuropeanUnion | Pork | 19242 | 19317 | 19746 | 20043 | 19773 | 19768 | 20015 |
| EuropeanUnion | Poultry | 6934 | 7359 | 7417 | 7312 | 7280 | 7596 | 7380 |
| Japan | Beef | 1585 | 1419 | 1319 | 1366 | 1182 | 1200 | 1173 |
| Japan | Pork | 2228 | 2268 | 2377 | 2373 | 2562 | 2507 | 2450 |
| Japan | Poultry | 1772 | 1797 | 1830 | 1841 | 1713 | 1880 | 1908 |
| Mexico | Beef | 2309 | 2341 | 2409 | 2308 | 2368 | 2419 | 2509 |
| Mexico | Pork | 1252 | 1298 | 1349 | 1423 | 1556 | 1556 | 1580 |
| Mexico | Poultry | 2163 | 2311 | 2424 | 2627 | 2713 | 2871 | 3005 |
| Russia | Beef | 2246 | 2400 | 2450 | 2378 | 2308 | 2503 | 2370 |
| Russia | Pork | 2019 | 2076 | 2453 | 2420 | 2337 | 2476 | 2637 |
| Russia | Poultry | 1320 | 1588 | 1697 | 1680 | 1675 | 2139 | 2382 |
| UnitedStates | Beef | 12502 | 12351 | 12737 | 12340 | 12667 | 12663 | 12830 |
| UnitedStates | Pork | 8455 | 8389 | 8685 | 8816 | 8817 | 8669 | 8640 |
| UnitedStates | Poultry | 11474 | 11558 | 12270 | 12540 | 13080 | 13430 | 13754 |
Exploratory data analyses (EDA)
In the following analysis, we will aim to perform an analysis of variance (ANOVA) to compare the meat consumption amounts between different countries and/or across time. Note that the data points for each country-meat type combination are from the various years. Typically, we would expect the amount not to change between the years (especially in this 7-year timespan). Even if it did, in assuming homoscedasticity, we are making the assumption that any increase or decrease is constant between countries. Applying the Fligner-Killeen test will help us decide if this assumption is valid. Look at the bar graphs listed below and note which of them seem to vary more than the others between the years.




Quantitative data analysis (QDA)
Open the SOCR ANOVA-Two Way applet (requires Java-enabled browser).
For the following analyses, we will focus on the data for Beef consumption. Let us test to see if different countries eat different amounts of beef. Usually when we compare a set of groups as this one, we would use a one-way ANOVA (comparing the seven countries). To run this test, open up the one-way ANOVA analysis in the SOCR Analyses Applet in a java-enabled browser. It should be the default setting when you open up the page:

Now, prepare your dataset (it will be the Beef table from the above summary tables). We will be treating the yearly results as being our sample’s data points, attempting to capture the overall population consumption. This seems reasonable; the average meet consumption of a country should not change that much in a seven-year timespan.
Once you have rearranged your dataset for use in the ANOVA applet (if you do not know what it should look like, try considering one of the SOCR ANOVA tutorials). It should look like this in the applet data screen:

Rename your column headers to define the independent and dependent variables.
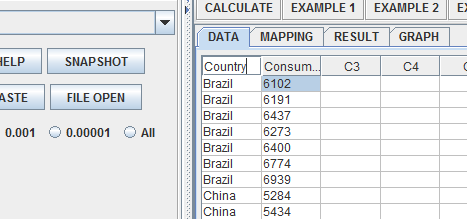
Click on the mapping tab, and assign your independent and dependent variables appropriately:

Set your precision to all, then click calculate:
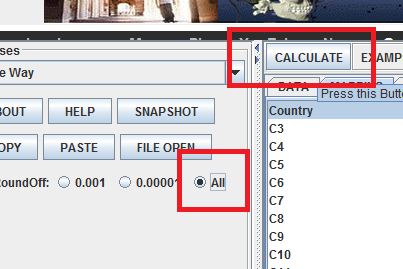
The following results should appear in the Results tab:
- Sample Size = 49
- Independent Variable = Country
- Dependent Variable = Consumption
- Results of One-Way Analysis of Variance:
- Standard 1-Way ANOVA Table
============================================================================================== VarianceSource DF RSS MSS F-Statistics P-value TreatmentEffect (B/w Groups) 6 668292765.4285718000 111382127.5714286400 898.8918351858 < 1E-15 Error 42 5204240.5714285690 123910.4897959183 Total: 48 673497006.0000004000 ==============================================================================================
- Model:
- Degrees of Freedom = 6
- Residual Sum of Squares = 668292765.4285718000
- Mean Square Error = 111382127.5714286400
- Error:
- Degrees of Freedom = 42
- Residual Sum of Squares = 5204240.5714285690
- Mean Square Error = 123910.4897959183
- Corrected Total:
- Degrees of Freedom = 48
- Residual Sum of Squares = 673497006.0000004000
- F-Value = 898.8918351858
- P-Value = < 1E-15
- R-Square = 0.9922728082
Conclusions
According to the results of the analysis, you will find that there is are significant main effects of locality (F(2, 106) = 18.39651, p < 0.001) and sex (F(1, 106) = 119.23809, p < 0.001) on shell width. The interaction between sex and locality is not significant on shell width (F (2,106) = 1.55056, p > 0.20). Post-hoc tests reveal that t-tests will reveal that there is a significant difference in width between male (M 7106.88136, SD = 247.06778) and female (M = 7578.03773, SD = 256.89806) snails shells (t (110) = 9.88846, p < 0.001). The 99.7% confidence interval for the difference is 471.15638 ± 157.08993. Note that this interval does not include 0 (a lack of difference between the means). There is also a significant difference in width between the snails collected at localities one and two, two and three, & one and three. We leave these analyses to you in the first practice problems
Based on these results, it would be possible to classify whether a Cocholotoma septemspirale is male or female, regardless of the locality it comes from (there is no interaction of the two effects); females have significantly taller shells. Limitations of the study include its correlational nature. One issue with the study, for example, is that age might be a confounding variable, if these snails are the type that grows throughout their lifecycle.
Practice problems
- Finish the post-hoc t-tests for the effect of locality on shell width.
- Complete an analysis similar to the one above, using one of the variables other than shell.h as -your dependent variable. See if that variable would be of use in classifying the snails.
- Complete a new analysis of this pain/neuroimaging data set. Use sex and disease group as independent variables. Choose for your dependent variable one of the brain volumes.
See also
References
- Che, Annie, Cui, Jenny, and Dinov, Ivo (2009). SOCR Analyses: Implementation and Demonstration of a New Graphical Statistics Educational Toolkit. JSS, Vol. 30, Issue 3, Apr 2009.
- Che, A, Cui, J, and Dinov, ID (2009) SOCR Analyses – an Instructional Java Web-based Statistical Analysis Toolkit, JOLT, 5(1), 1-19, March 2009.
- Dinov, ID. Statistics Online Computational Resource, Journal of Statistical Software, Vol. 16, No. 1, 1-16, October 2006.
- Reichenbach F, Baur H, Neubert E (2012) Sexual dimorphism in shells of Cochlostoma septemspirale (Caenogastropoda, Cyclophoroidea, Diplommatinidae, Cochlostomatinae). ZooKeys 208: 1-16. doi:10.3897/zookeys.208.2869
- Baur H, Reichenbach F, Neubert E (2012) Data from: Sexual dimorphism in shells of Cochlostoma septemspirale (Caenogastropoda, Cyclophoroidea, Diplommatinidae, Cochlostomatinae). Dryad Digital Repository. doi:10.5061/dryad.ns7v7
Translate this page: